class: center, top, title-slide .title[ # Statistical Inference ] .subtitle[ ## IQA Lecture 3 ] .author[ ### Charles Lanfear ] .date[ ### 29 Oct 2025<br>Updated: 28 Oct 2025 ] --- # Today * Two notes * Course structure * Research portfolio * Code * `ggplot2` * More tables * `{broom}` for model output * Stats * Chi-square test * Simple OLS regression --- class: inverse # Some Review --- # London Crime Let's load another example dataset: Real Metropolitan Police recorded crime in the 32 London boroughs (no City of London) in 2021 ``` r library(tidyverse) metro_2021 <- read_csv("https://clanfear.github.io/ioc_iqa/_data/metro_2021.csv") ``` I obtained these from `data.police.uk`—it is easy to get your own for nearly anywhere in the UK, and in any recent period. I also linked them to data on deprivation and subregions, which would take more work!<sup>1</sup> .footnote[ [1] We'll actually make these in CRMW next term! ] --- # London Crime ``` r glimpse(metro_2021) ``` ``` ## Rows: 384 ## Columns: 10 ## $ borough <chr> "Barking and Dagenham", "Bark… ## $ month <date> 2021-01-01, 2021-02-01, 2021… ## $ antisocial_behaviour <dbl> 688, 585, 600, 531, 469, 507,… ## $ burglary <dbl> 87, 105, 81, 85, 106, 90, 75,… ## $ robbery <dbl> 30, 50, 49, 40, 47, 46, 42, 3… ## $ violence_and_sexual_offences <dbl> 469, 449, 561, 564, 589, 617,… ## $ subregion <chr> "East", "East", "East", "East… ## $ deprivation <chr> "High", "High", "High", "High… ## $ pop <dbl> 221495, 221495, 221495, 22149… ## $ area <dbl> 36.1, 36.1, 36.1, 36.1, 36.1,… ``` --- # `distinct()` These data have monthly observations for each borough. If we wanted to look at only unique observations of boroughs, we can use `distinct()` ``` r metro_2021 |> distinct(borough) |> head() ``` ``` ## # A tibble: 6 × 1 ## borough ## <chr> ## 1 Barking and Dagenham ## 2 Barnet ## 3 Bexley ## 4 Brent ## 5 Bromley ## 6 Camden ``` --- # `summarize()` Just as a quick check, let's see how many boroughs and months are in the data ``` r metro_2021 |> summarize(n_boroughs = n_distinct(borough), n_months = n_distinct(month), sample_size = n()) ``` ``` ## # A tibble: 1 × 3 ## n_boroughs n_months sample_size ## <int> <int> <int> ## 1 32 12 384 ``` Looks like every borough is seen in 12 months ``` r 32 * 12 ``` ``` ## [1] 384 ``` --- # Another Check A different way to see if all boroughs are observed 12 times is to count observations of each borough twice ``` r metro_2021 |> count(borough) |> head(3) ``` ``` ## # A tibble: 3 × 2 ## borough n ## <chr> <int> ## 1 Barking and Dagenham 12 ## 2 Barnet 12 ## 3 Bexley 12 ``` -- Okay, then count how many of each value of `n` ``` r *metro_2021 |> count(borough) |> count(n) # This makes a warning! ``` ``` ## # A tibble: 1 × 2 ## n nn ## <int> <int> ## 1 12 32 ``` Every value of `n` was 12, so there were 32 values of 12. --- # Saving a Subset If we think a particular subset will be used repeatedly, we can save it and give it a name like any other object: ``` r westminster <- metro_2021 |> filter(borough == "Westminster") head(westminster, 4) ``` ``` ## # A tibble: 4 × 10 ## borough month antisocial_behaviour burglary robbery ## <chr> <date> <dbl> <dbl> <dbl> ## 1 Westminster 2021-01-01 1501 150 64 ## 2 Westminster 2021-02-01 1326 150 51 ## 3 Westminster 2021-03-01 1474 189 76 ## 4 Westminster 2021-04-01 1396 155 126 ## # ℹ 5 more variables: violence_and_sexual_offences <dbl>, ## # subregion <chr>, deprivation <chr>, pop <dbl>, area <dbl> ``` -- .text-center[ *And now for something completely different* ] --- class: inverse # Plotting  --- ## Base R Plots from 2 Weeks Ago .pull-left[ ``` r plot(robbery ~ month, data = westminster, xlab = "Month", ylab = "Robbery", main = "Robbery in Westminster", col = "red", cex.lab = 1.5, cex.main= 1.5, pch = 16) ``` ] .pull-right[  ] --- # `ggplot2` An alternative way of plotting many prefer (myself included) uses the `ggplot2` package in R, which is part of the `tidyverse`. ``` r library(ggplot2) ``` The core idea underlying this package is the [**layered grammar of graphics**](https://doi.org/10.1198/jcgs.2009.07098): we can break up elements of a plot into pieces and combine them. --- ## Robberies in `ggplot` .pull-left[ ``` r ggplot(data = westminster, aes(x = month, y = robbery)) + geom_point() ``` ] .pull-right[  ] .footnote[ *Weird syntax and an ugly plot—what's going on here?* ] --- # Structure of a ggplot `ggplot2` graphics objects consist of two primary components: -- **Layers**, the components of a graph. * We *add* layers to a `ggplot2` object using `+`. * This includes lines, shapes, and text. -- **Aesthetics**, which determine how the layers appear. * We *set* aesthetics using *arguments* (e.g. `color="red"`) inside layer functions. * This includes locations, colors, and sizes. * Aesthetics also determine how data *map* to appearances. --- # Layers **Layers** are the components of the graph, such as: * `ggplot()`: The initial plot canvas * `geom_point()`: Scatterplot points * `geom_line()`: Lines between data points * `labs()`: Plot title and axis labels * `facet_wrap()`: Divides plot into a sequence of panels * `facet_grid()`: Divides plot into a grid of plot panels * `theme_bw()`: Replace default gray background with black-and-white Layers are separated by a `+` sign. --- # Aesthetics **Aesthetics** control the appearance of the layers: * `x`, `y`: `\(x\)` and `\(y\)` coordinate values to use * `color`: Color of elements based on some data value * `group`: Describe which data points are grouped together (often used with lines) * `size`: Size of points/lines based on some data value * `alpha`: Transparency based on some data value --- ## Aesthetics: Setting vs. mapping Layers take arguments to control their appearance, such as point/line colors or transparency (`alpha` between 0 and 1). -- * Arguments like `color`, `size`, and `fill` can be used directly on layers (**setting aesthetics**), e.g. `geom_point(color = "red")`. These *don't depend on the data*. -- * Arguments inside `aes()` (**mapping aesthetics**) will *depend on the data*, e.g. `geom_point(aes(color = continent))`. -- * `aes()` in the `ggplot()` layer gives overall aesthetics to use in other layers, but can be changed on individual layers (including switching `x` or `y` to different variables) -- This may seem pedantic, but precise language makes searching for help easier. -- Now let's see all this jargon in action. --- # Westminster Robbery ### 1: Base Plot .pull-left[ .text-85[ ``` r *ggplot(data = westminster, * aes(x = month, * y = robbery)) ``` ] ] .pull-right[  ] .footnote[Initialize the plot with `ggplot()` and `x` and `y` aesthetics **mapped** to variables.] --- # Westminster Robbery ### 2: Scatterplot .pull-left[ .text-85[ ``` r ggplot(data = westminster, aes(x = month, y = robbery)) + * geom_point() ``` ] ] .pull-right[  ] .footnote[Add a scatterplot **layer**] --- # Westminster Robbery ### 3: Point Color and Size .pull-left[ .text-85[ ``` r ggplot(data = westminster, aes(x = month, y = robbery)) + geom_point(color = "red", * size = 3) ``` ] ] .pull-right[  ] .footnote[**Set** aesthetics to make the points large and red] --- # Westminster Robbery ### 4: X-Axis Label .pull-left[ .text-85[ ``` r ggplot(data = westminster, aes(x = month, y = robbery)) + geom_point(color = "red", size = 3) + * labs(x = "Month") ``` ] ] .pull-right[  ] .footnote[Add a label layer to capitalize the x-axis label] --- # Westminster Robbery ### 5: Y-Axis Label .pull-left[ .text-85[ ``` r ggplot(data = westminster, aes(x = month, y = robbery)) + geom_point(color = "red", size = 3) + labs(x = "Month", * y = "Robbery") ``` ] ] .pull-right[  ] .footnote[Add text to clean up the y-axis label] --- # Westminster Robbery ### 6: Title .pull-left[ .text-85[ ``` r ggplot(data = westminster, aes(x = month, y = robbery)) + geom_point(color = "red", size = 3) + labs(x = "Month", y = "Robbery", title = * "Robbery in Westminster") ``` ] ] .pull-right[  ] .footnote[Add a title] --- # Westminster Robbery ### 7: Theme .pull-left[ .text-85[ ``` r ggplot(data = westminster, aes(x = month, y = robbery)) + geom_point(color = "red", size = 3) + labs(x = "Month", y = "Robbery", title = "Robbery in Westminster") + * theme_bw() ``` ] ] .pull-right[  ] .footnote[Pick a basic theme with a new layer] --- # Westminster Robbery ### 8: Text Size .pull-left[ .text-85[ ``` r ggplot(data = westminster, aes(x = month, y = robbery)) + geom_point(color = "red", size = 3) + labs(x = "Month", y = "Robbery", title = "Robbery in Westminster") + * theme_bw(base_size = 18) ``` ] ] .pull-right[  ] .footnote[Increase the base text size] --- # Plotting All Boroughs We have a plot we like for Westminster... ... but what if we want *all the boroughs*? --- # Plotting All Boroughs ### 1: A Mess! .pull-left[ .text-85[ ``` r ggplot(data = metro_2021, aes(x = month, y = robbery)) + geom_point(color = "red", size = 3) + labs(x = "Month", y = "Robbery", title = "Robbery in London") + theme_bw(base_size = 18) ``` ] ] .pull-right[  ] .footnote[We can't tell boroughs apart! Maybe we could follow *lines*?] --- # Plotting All Boroughs ### 2: Lines .pull-left[ .text-85[ ``` r ggplot(data = metro_2021, aes(x = month, y = robbery)) + geom_line(color = "red", * linewidth = 3) + labs(x = "Month", y = "Robbery", title = "Robbery in London") + theme_bw(base_size = 18) ``` ] ] .pull-right[  ] .footnote[`ggplot2` doesn't know how to connect the lines!] --- # Plotting All Boroughs ### 3: Grouping .pull-left[ .text-85[ ``` r ggplot(data = metro_2021, aes(x = month, y = robbery, group = borough)) + geom_line(color = "red", linewidth = 3) + labs(x = "Month", y = "Robbery", title = "Robbery in London")+ theme_bw(base_size = 18) ``` ] ] .pull-right[  ] .footnote[That looks more reasonable... but the lines are too thick!] --- # Plotting All Boroughs ### 4: Size .pull-left[ .text-85[ ``` r ggplot(data = metro_2021, aes(x = month, y = robbery, group = borough)) + * geom_line(color = "red") + labs(x = "Month", y = "Robbery", title = "Robbery in London") + theme_bw(base_size = 18) ``` ] ] .pull-right[  ] .footnote[Much better... but maybe we can do highlight subregions?] --- # Plotting All Boroughs ### 5: Color .pull-left[ .text-85[ ``` r ggplot(data = metro_2021, aes(x = month, y = robbery, group = borough, * color = subregion)) + geom_line() + labs(x = "Month", y = "Robbery", title = "Robbery in London") + theme_bw(base_size = 18) ``` ] ] .pull-right[  ] .footnote[Hmmm, hard to make out... why not separate subregions completely?] --- # Plotting All Boroughs ### 6: Facets .pull-left[ .text-85[ ``` r ggplot(data = metro_2021, aes(x = month, y = robbery, group = borough, color = subregion)) + geom_line() + * facet_wrap(~subregion) + labs(x = "Month", y = "Robbery", title = "Robbery in London")+ theme_bw(base_size = 18) ``` ] ] .pull-right[  ] .footnote[Looking good, but the legend is redundant!] --- # Plotting All Boroughs ### 7: Legend .pull-left[ .text-85[ ``` r ggplot(data = metro_2021, aes(x = month, y = robbery, group = borough, color = subregion)) + geom_line() + facet_wrap(~subregion) + labs(x = "Month", y = "Robbery", title = "Robbery in London") + theme_bw(base_size = 18) + * theme(legend.position = "none") ``` ] ] .pull-right[  ] .footnote[The x-axis labels are overlapping!] --- # Plotting All Boroughs ### 8: Axis Labels .pull-left[ .text-85[ ``` r ggplot(data = metro_2021, aes(x = month, y = robbery, group = borough, color = subregion)) + geom_line() + * scale_x_date(date_labels = "%b") + facet_wrap(~subregion) + labs(x = "Month", y = "Robbery", title = "Robbery in London") + theme_bw(base_size = 18) + theme(legend.position = "none") ``` You can look at the help file `?strptime` to find these `%` codes for date formatting ] ] .pull-right[  ] .footnote[Not bad!] --- # Enough for now .pull-left-60[ Don't worry about understanding it all now—it is complicated! ... but it is worth learning because it can produce publication ready plots We'll return to `ggplot2` at least for a bit in every future lecture ] .pull-right-40[  ] .footnote[ .pull-left[ ‍Source: [Lanfear et al. (2023) "Inequalities in Exposure to Firearm Violence by Race, Sex, and Birth Cohort From Childhood to Age 40 Years, 1995-2021"](https://jamanetwork.com/journals/jamanetworkopen/fullarticle/2804655) ] ] --- background-color: white # Bonus map  .footnote[ ‍Source: [Lanfear & Kirk (2024) "The Promise and Perils of the Sharing Economy: The Impact of Airbnb Lettings on Crime"](https://doi.org/10.1111/1745-9125.12383) ] --- class: inverse # Book Recommendation .pull-left[  ] .pull-right[ * Targeted at social scientists without technical backgrounds * Teaches good visualization principles * Uses R, `ggplot2`, and `tidyverse` * Free online version: * [https://socviz.co/](https://socviz.co/) * Affordable in print ] --- class: inverse # Statistical Inference  --- # `communities` Data Let's return to the `communities` data we've used before—making sure to fix the ordinal variables again .text-85[ ``` r communities <- read_csv("https://clanfear.github.io/ioc_iqa/_data/communities.csv") |> mutate(disadvantage = factor(disadvantage, levels = c("Low", "Medium", "High")), incarceration = factor(incarceration, levels = c("Low", "Medium", "High"))) glimpse(communities) ``` ``` ## Rows: 300 ## Columns: 5 ## $ area <chr> "Urban", "Urban", "Rural", "Rural", "Urban",… ## $ pop_density <dbl> 18.166008, 21.356727, 10.023975, 14.926138, … ## $ crime_rate <dbl> 28.63123, 58.60800, 14.19840, 22.29075, 79.7… ## $ disadvantage <fct> Low, Medium, Low, Medium, High, High, Low, L… ## $ incarceration <fct> High, Medium, High, Medium, High, High, High… ``` ] --- # Cross-Tab Say we tabulate our data and see this association in our sample: ``` r library(janitor) # For the tabyl function communities |> tabyl(disadvantage, area) |> adorn_title() ``` ``` ## area ## disadvantage Rural Urban ## Low 63 39 ## Medium 49 48 ## High 41 60 ``` It looks like disadvantage has different patterns in rural and urban areas -- We want to know if there's a real relationship in the population How do we figure this out? --- # Chi-square Test `janitor` adds a `chisq.test()` function that works on its special tables ``` r communities |> tabyl(disadvantage, area) |> chisq.test() ``` ``` ## ## Pearson's Chi-squared test ## ## data: tabyl(communities, disadvantage, area) ## X-squared = 9.1153, df = 2, p-value = 0.01049 ``` -- This is the same result we saw in CRM yesterday: There appears to be an association. -- If you're curious how p-values are calculated, they come from cumulative probability distributions—which R has built in! ``` r 1 - pchisq(9.1153, df = 2) ``` ``` ## [1] 0.01048667 ``` .pull-right[ .footnote[We'll mess with this a bit more another day!] ] --- ## How unlikely is that? .pull-left[ .text-85[ ``` r tibble(x = seq(0, 15, 0.1), density = dchisq(x, df = 2)) |> ggplot(aes(x = x, y = density)) + geom_line() + coord_cartesian(expand = FALSE) + geom_vline(xintercept = 9.1153, color = "red") ``` ] ] .pull-right[  ] That is really unlikely to occur by chance: *If there were no real association in the population, only 1.04% of all possible random samples would have an association this strong* --- # A Prettier Table There are many ways to make nicer looking tables in R For very simple tables using dataframes, I use `kable()` in `{knitr}` ``` r communities |> tabyl(disadvantage, area) |> adorn_title() |> * knitr::kable() # Access a package function without loading it ``` | |area | | |:------------|:-----|:-----| |disadvantage |Rural |Urban | |Low |63 |39 | |Medium |49 |48 | |High |41 |60 | .footnote[ Yours won't have the colored rows—that's from my website theme! ] --- # More Complex If you want to make a publication ready table that will work in any output format, such as PDF, Word, or HTML, [your best bet is `{flextable}`](https://ardata-fr.github.io/flextable-book/) ``` r library(flextable) communities |> rename(Disadvantage = disadvantage) |> tabyl(Disadvantage, area) |> flextable() |> add_header_lines("Table 1. Disadvantage by area") ``` <div class="tabwid"><style>.cl-8c40a4fc{}.cl-8c38b6e8{font-family:'Arial';font-size:11pt;font-weight:normal;font-style:normal;text-decoration:none;color:rgba(0, 0, 0, 1.00);background-color:transparent;}.cl-8c3bbd5c{margin:0;text-align:left;border-bottom: 0 solid rgba(0, 0, 0, 1.00);border-top: 0 solid rgba(0, 0, 0, 1.00);border-left: 0 solid rgba(0, 0, 0, 1.00);border-right: 0 solid rgba(0, 0, 0, 1.00);padding-bottom:5pt;padding-top:5pt;padding-left:5pt;padding-right:5pt;line-height: 1;background-color:transparent;}.cl-8c3bbd70{margin:0;text-align:right;border-bottom: 0 solid rgba(0, 0, 0, 1.00);border-top: 0 solid rgba(0, 0, 0, 1.00);border-left: 0 solid rgba(0, 0, 0, 1.00);border-right: 0 solid rgba(0, 0, 0, 1.00);padding-bottom:5pt;padding-top:5pt;padding-left:5pt;padding-right:5pt;line-height: 1;background-color:transparent;}.cl-8c3bd4e0{width:0.75in;background-color:transparent;vertical-align: middle;border-bottom: 1.5pt solid rgba(102, 102, 102, 1.00);border-top: 1.5pt solid rgba(102, 102, 102, 1.00);border-left: 0 solid rgba(0, 0, 0, 1.00);border-right: 0 solid rgba(0, 0, 0, 1.00);margin-bottom:0;margin-top:0;margin-left:0;margin-right:0;}.cl-8c3bd4e1{width:0.75in;background-color:transparent;vertical-align: middle;border-bottom: 1.5pt solid rgba(102, 102, 102, 1.00);border-top: 1.5pt solid rgba(102, 102, 102, 1.00);border-left: 0 solid rgba(0, 0, 0, 1.00);border-right: 0 solid rgba(0, 0, 0, 1.00);margin-bottom:0;margin-top:0;margin-left:0;margin-right:0;}.cl-8c3bd4ea{width:0.75in;background-color:transparent;vertical-align: middle;border-bottom: 0 solid rgba(0, 0, 0, 1.00);border-top: 0 solid rgba(0, 0, 0, 1.00);border-left: 0 solid rgba(0, 0, 0, 1.00);border-right: 0 solid rgba(0, 0, 0, 1.00);margin-bottom:0;margin-top:0;margin-left:0;margin-right:0;}.cl-8c3bd4f4{width:0.75in;background-color:transparent;vertical-align: middle;border-bottom: 0 solid rgba(0, 0, 0, 1.00);border-top: 0 solid rgba(0, 0, 0, 1.00);border-left: 0 solid rgba(0, 0, 0, 1.00);border-right: 0 solid rgba(0, 0, 0, 1.00);margin-bottom:0;margin-top:0;margin-left:0;margin-right:0;}.cl-8c3bd4f5{width:0.75in;background-color:transparent;vertical-align: middle;border-bottom: 1.5pt solid rgba(102, 102, 102, 1.00);border-top: 0 solid rgba(0, 0, 0, 1.00);border-left: 0 solid rgba(0, 0, 0, 1.00);border-right: 0 solid rgba(0, 0, 0, 1.00);margin-bottom:0;margin-top:0;margin-left:0;margin-right:0;}.cl-8c3bd4f6{width:0.75in;background-color:transparent;vertical-align: middle;border-bottom: 1.5pt solid rgba(102, 102, 102, 1.00);border-top: 0 solid rgba(0, 0, 0, 1.00);border-left: 0 solid rgba(0, 0, 0, 1.00);border-right: 0 solid rgba(0, 0, 0, 1.00);margin-bottom:0;margin-top:0;margin-left:0;margin-right:0;}</style><table data-quarto-disable-processing='true' class='cl-8c40a4fc'><thead><tr style="overflow-wrap:break-word;"><th colspan="3"class="cl-8c3bd4e0"><p class="cl-8c3bbd5c"><span class="cl-8c38b6e8">Table 1. Disadvantage by area</span></p></th></tr><tr style="overflow-wrap:break-word;"><th class="cl-8c3bd4e0"><p class="cl-8c3bbd5c"><span class="cl-8c38b6e8">Disadvantage</span></p></th><th class="cl-8c3bd4e1"><p class="cl-8c3bbd70"><span class="cl-8c38b6e8">Rural</span></p></th><th class="cl-8c3bd4e1"><p class="cl-8c3bbd70"><span class="cl-8c38b6e8">Urban</span></p></th></tr></thead><tbody><tr style="overflow-wrap:break-word;"><td class="cl-8c3bd4ea"><p class="cl-8c3bbd5c"><span class="cl-8c38b6e8">Low</span></p></td><td class="cl-8c3bd4f4"><p class="cl-8c3bbd70"><span class="cl-8c38b6e8">63</span></p></td><td class="cl-8c3bd4f4"><p class="cl-8c3bbd70"><span class="cl-8c38b6e8">39</span></p></td></tr><tr style="overflow-wrap:break-word;"><td class="cl-8c3bd4ea"><p class="cl-8c3bbd5c"><span class="cl-8c38b6e8">Medium</span></p></td><td class="cl-8c3bd4f4"><p class="cl-8c3bbd70"><span class="cl-8c38b6e8">49</span></p></td><td class="cl-8c3bd4f4"><p class="cl-8c3bbd70"><span class="cl-8c38b6e8">48</span></p></td></tr><tr style="overflow-wrap:break-word;"><td class="cl-8c3bd4f5"><p class="cl-8c3bbd5c"><span class="cl-8c38b6e8">High</span></p></td><td class="cl-8c3bd4f6"><p class="cl-8c3bbd70"><span class="cl-8c38b6e8">41</span></p></td><td class="cl-8c3bd4f6"><p class="cl-8c3bbd70"><span class="cl-8c38b6e8">60</span></p></td></tr></tbody></table></div> It can be complicated, but it can do just about anything... --- # `{flextable}` is... flexible  .footnote[Source: [Flextable Gallery](https://www.ardata.fr/en/flextable-gallery/2021-01-12-mario-kart/)] --- ### I suppose you can use it for articles too 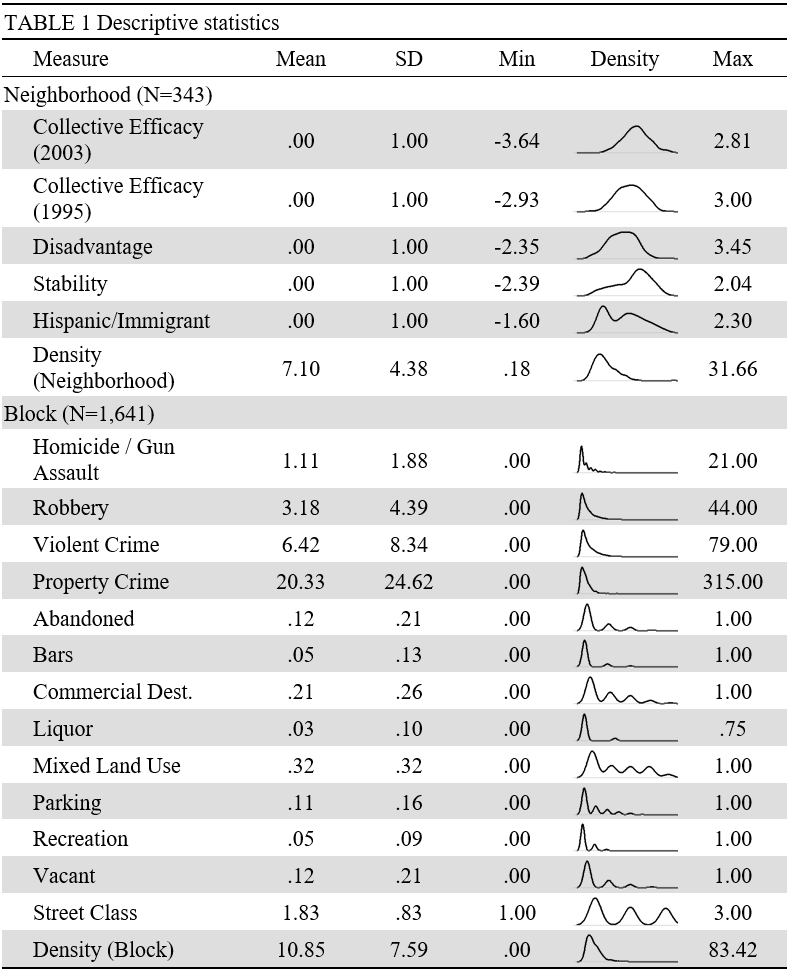 .footnote[Source: [Lanfear (2022) "Collective efficacy and the built environment"](https://doi.org/10.1111/1745-9125.12304)] --- # Bar Plots Aside from tables, you may want to plot categorical data .pull-left[ ``` r ggplot(communities, aes(x = disadvantage)) + geom_bar() ``` `geom_bar()` tabulates counts of values of categorical variables and makes a bar plot ] .pull-right[  ] --- # Filled Bar Plots Adding a `fill` aesthetic will color the bars by a second variable .pull-left[ ``` r ggplot(communities, aes(x = disadvantage, fill = area)) + geom_bar() ``` ] .pull-right[  ] --- # Dodgy Bar Plots Adding `position = "dodge"` in `geom_bar()` will unstack them .pull-left[ ``` r ggplot(communities, aes(x = disadvantage, fill = area)) + geom_bar(position = "dodge") ``` ] .pull-right[  ] --- class: inverse # Linear Regression <video controls autoplay> <source src="img/parks-rec-webm.mp4" type="video/mp4"> </video> .footnote[Credit: [Edmund Helmur](https://twitter.com/theotheredmund/status/1349453230762196992?s=20&t=bBDscDYt2was9ABePCcZhQ)] --- # `lm()` We can run an OLS linear model in R using `lm()` -- It takes a **formula** as the first argument, e.g. `y ~ x` ``` r cr_pd_lm <- lm(crime_rate ~ pop_density, data = communities) cr_pd_lm ``` ``` ## ## Call: ## lm(formula = crime_rate ~ pop_density, data = communities) ## ## Coefficients: ## (Intercept) pop_density ## -20.777 3.094 ``` By default, it won't produce much output—you probably wanted significance tests! --- # `summary()` To get detailed output, use `summary()` on the output from `lm()` .text-85[ ``` r summary(cr_pd_lm) ``` ``` ## ## Call: ## lm(formula = crime_rate ~ pop_density, data = communities) ## ## Residuals: ## Min 1Q Median 3Q Max ## -35.171 -7.423 -0.983 5.584 55.867 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) -20.7769 2.0909 -9.937 <2e-16 *** ## pop_density 3.0939 0.1314 23.544 <2e-16 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 12.9 on 298 degrees of freedom ## Multiple R-squared: 0.6504, Adjusted R-squared: 0.6492 ## F-statistic: 554.3 on 1 and 298 DF, p-value: < 2.2e-16 ``` ] --- # `lm()` Output is a List `lm()` produces a lot more information than what is shown by `summary()` however. We can see the structure of `lm()` output using `str()`: .text-85[ ``` r str(cr_pd_lm, list.len=6) # Truncating the output a bit ``` ``` ## List of 12 ## $ coefficients : Named num [1:2] -20.78 3.09 ## ..- attr(*, "names")= chr [1:2] "(Intercept)" "pop_density" ## $ residuals : Named num [1:300] -6.8 13.31 3.96 -3.11 32.83 ... ## ..- attr(*, "names")= chr [1:300] "1" "2" "3" "4" ... ## $ effects : Named num [1:300] -436.83 -303.74 4.99 -2.73 32.31 ... ## ..- attr(*, "names")= chr [1:300] "(Intercept)" "pop_density" "" "" ... ## $ rank : int 2 ## $ fitted.values: Named num [1:300] 35.4 45.3 10.2 25.4 46.9 ... ## ..- attr(*, "names")= chr [1:300] "1" "2" "3" "4" ... ## $ assign : int [1:2] 0 1 ## [list output truncated] ## - attr(*, "class")= chr "lm" ``` ] `lm()` actually has an enormous quantity of output! This is a type of object called a list. --- # Digging Into a List We can access parts of lm() output using $ like with dataframe names: ``` r cr_pd_lm$coefficients ``` ``` ## (Intercept) pop_density ## -20.776945 3.093886 ``` We can also do this with summary(), which provides additional statistics: ``` r summary(cr_pd_lm)$coefficients ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) -20.776945 2.0908640 -9.937014 2.769101e-20 ## pop_density 3.093886 0.1314093 23.543883 5.701147e-70 ``` --- # `broom::tidy()` A quick way to get model output into an easy-to-use format is with the `{broom}` package's `tidy()` function ``` r library(broom) tidy(cr_pd_lm) ``` ``` ## # A tibble: 2 × 5 ## term estimate std.error statistic p.value ## <chr> <dbl> <dbl> <dbl> <dbl> ## 1 (Intercept) -20.8 2.09 -9.94 2.77e-20 ## 2 pop_density 3.09 0.131 23.5 5.70e-70 ``` -- `tidy()` turns model output into a dataframe—now you can use `dplyr` with it! ``` r tidy(cr_pd_lm) |> filter(term == "pop_density") ``` ``` ## # A tibble: 1 × 5 ## term estimate std.error statistic p.value ## <chr> <dbl> <dbl> <dbl> <dbl> ## 1 pop_density 3.09 0.131 23.5 5.70e-70 ``` --- # `broom::augment()` `{broom}` can also add regression results back to your original data using `augment()` This includes **predicted values** and **residuals** ``` r cr_pd_lm |> augment() |> glimpse() ``` ``` ## Rows: 300 ## Columns: 8 ## $ crime_rate <dbl> 28.63123, 58.60800, 14.19840, 22.29075, 79.772… ## $ pop_density <dbl> 18.166008, 21.356727, 10.023975, 14.926138, 21… ## $ .fitted <dbl> 35.426612, 45.298331, 10.236091, 25.402822, 46… ## $ .resid <dbl> -6.7953821, 13.3096695, 3.9623093, -3.1120717,… ## $ .hat <dbl> 0.004462361, 0.007702732, 0.005767089, 0.00333… ## $ .sigma <dbl> 12.91673, 12.89950, 12.92072, 12.92151, 12.780… ## $ .cooksd <dbl> 6.245907e-04, 4.163074e-03, 2.751664e-04, 9.76… ## $ .std.resid <dbl> -0.52790919, 1.03566799, 0.30801969, -0.241628… ``` We'll use this later to do model diagnostics! --- # Plotting Regression Lines `geom_smooth()` is used to add conditional mean lines to plots—the regression line is just one type of these! .pull-left[ .text-85[ ``` r communities |> ggplot(aes(x = pop_density, y = crime_rate)) + geom_point(alpha = 0.5) + labs(x = "Population Density", y = "Crime Rate") + geom_smooth(method = "lm", formula = "y ~ x") + theme_minimal(base_size = 16) ``` By default it includes a confidence interval along the line This is the CI for the mean *at each level of `x`* ] ] .pull-right[  ] --- # More Lines If you group your data, you can draw a regression line for each separately—equivalent to running a separate model on each group .pull-left[ .text-85[ ``` r communities |> ggplot(aes(x = pop_density, y = crime_rate, group = area, color = area)) + geom_point(alpha = 0.5) + labs(x = "Population Density", y = "Crime Rate", color = "Area") + geom_smooth(method = "lm", formula = "y ~ x") + theme_minimal(base_size = 16) ``` ] ] .pull-right[  ] --- class: inverse # Wrap-Up * That was a lot this week! * Take the week off on this course—focus on CRM and your other courses * Next week will have an assignment handed out * Most of the rest of the course is figuring out how to apply all this * Plus learning some more programming tools!