class: center, top, title-slide # R Visualization Workshop ## Plotting with <code>ggplot2</code> ### Chuck Lanfear ### May 4, 2019<br>Updated: May 4, 2019 --- class: inverse # Setup and Data --- # Gapminder Data We'll be working with data from Hans Rosling's [Gapminder](http://www.gapminder.org) project. An excerpt of these data can be accessed through an R package called `gapminder`, cleaned and assembled by Jenny Bryan at UBC. -- In the console: `install.packages("gapminder")` Load the package and data: ```r library(gapminder) ``` --- # Check Out Gapminder The data frame we will work with is called `gapminder`, available once you have loaded the package. Let's see its structure: .small[ ```r str(gapminder) ``` ``` ## Classes 'tbl_df', 'tbl' and 'data.frame': 1704 obs. of 6 variables: ## $ country : Factor w/ 142 levels "Afghanistan",..: 1 1 1 1 1 1 1 1 1 1 ... ## $ continent: Factor w/ 5 levels "Africa","Americas",..: 3 3 3 3 3 3 3 3 3 3 ... ## $ year : int 1952 1957 1962 1967 1972 1977 1982 1987 1992 1997 ... ## $ lifeExp : num 28.8 30.3 32 34 36.1 ... ## $ pop : int 8425333 9240934 10267083 11537966 13079460 14880372 12881816 13867957 16317921 22227415 ... ## $ gdpPercap: num 779 821 853 836 740 ... ``` ] --- # What's Interesting Here? * **Factor** variables `country` and `continent` + Factors are categorical data with an underlying numeric representation + We'll spend a lot of time on factors later! -- * Many observations: `\(n=1704\)` rows -- * A nested/hierarchical structure: `year` in `country` in `continent` + These are panel data! --- # Installing Tidyverse We'll want to be able to slice up this data frame into subsets (e.g. just the rows for Afghanistan, just the rows for 1997). We will use a package called `dplyr` to do this neatly. If you're unfamiliar, see my [previous workshop](https://clanfear.github.io/Intermediate_R_Workshop/)! `dplyr` is part of the [tidyverse](http://tidyverse.org/) family of R packages. -- If you have not already installed the tidyverse, type, in the console: `install.packages("tidyverse")` -- This will install a *large* number of R packages we will use throughout the term, including `dplyr` and `ggplot2`. `dplyr` is a very useful and powerful package that we will talk more about soon, but today we're mostly going to use it for subsetting data. --- # Subsetting Example For some examples to follow, I'd like to subset down only to data in China. ```r library(dplyr) China <- gapminder %>% filter(country == "China") head(China) ``` ``` ## # A tibble: 6 x 6 ## country continent year lifeExp pop gdpPercap ## <fct> <fct> <int> <dbl> <int> <dbl> ## 1 China Asia 1952 44 556263527 400. ## 2 China Asia 1957 50.5 637408000 576. ## 3 China Asia 1962 44.5 665770000 488. ## 4 China Asia 1967 58.4 754550000 613. ## 5 China Asia 1972 63.1 862030000 677. ## 6 China Asia 1977 64.0 943455000 741. ``` .footnote[If anything done here is unclear, let me know!] --- class: inverse # `ggplot2` .center[ <img src="img/ggplot2unit.jpg" style="width: 60%;"/> ] --- ## Base R Plots .pull-left[ .small[ ```r plot(lifeExp ~ year, data = China, xlab = "Year", ylab = "Life expectancy", main = "Life expectancy in China", col = "red", cex.lab = 1.5, cex.main= 1.5, pch = 16) ``` ] ] .pull-right[ 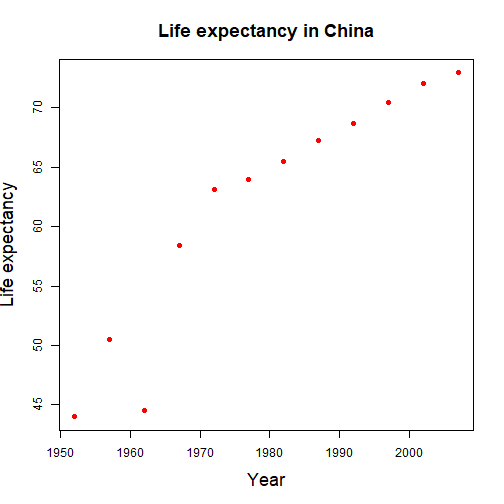<!-- --> ] --- # `ggplot2` An alternative way of plotting many prefer (myself included)<sup>1</sup> uses the `ggplot2` package in R, which is part of the `tidyverse`. .footnote[[1] [Though this is not without debate](http://simplystatistics.org/2016/02/11/why-i-dont-use-ggplot2/)] ```r library(ggplot2) ``` The core idea underlying this package is the [**layered grammar of graphics**](https://doi.org/10.1198/jcgs.2009.07098): we can break up elements of a plot into pieces and combine them. --- ## Chinese Life Expectancy in `ggplot` .pull-left[ .small[ ```r ggplot(data = China, aes(x = year, y = lifeExp)) + geom_point(color = "red", size = 3) + xlab("Year") + ylab("Life expectancy") + ggtitle("Life expectancy in China") + * theme_bw(base_size=18) ``` ] ] .pull-right[ <!-- --> ] --- # Structure of a ggplot `ggplot2` graphics objects consist of two primary components: -- 1. **Layers**, the components of a graph. * We *add* layers to a `ggplot2` object using `+`. * This includes lines, shapes, and text. -- 2. **Aesthetics**, which determine how the layers appear. * We *set* aesthetics using *arguments* (e.g. `color="red"`) inside layer functions. * This includes locations, colors, and sizes. * Aesthetics also determine how data *map* to appearances. --- # Layers **Layers** are the components of the graph, such as: * `ggplot()`: initializes `ggplot2` object, specifies input data * `geom_point()`: layer of scatterplot points * `geom_line()`: layer of lines * `ggtitle()`, `xlab()`, `ylab()`: layers of labels * `facet_wrap()`: layer creating separate panels stratified by some factor wrapping around * `facet_grid()`: same idea, but can split by two variables along rows and columns (e.g. `facet_grid(gender ~ age_group)`) * `theme_bw()`: replace default gray background with black-and-white Layers are separated by a `+` sign. For clarity, I usually put each layer on a new line, unless it takes few or no arguments (e.g. `xlab()`, `ylab()`, `theme_bw()`). --- # Aesthetics **Aesthetics** control the appearance of the layers: * `x`, `y`: `\(x\)` and `\(y\)` coordinate values to use * `color`: set color of elements based on some data value * `group`: describe which points are conceptually grouped together for the plot (often used with lines) * `size`: set size of points/lines based on some data value * `alpha`: set transparency based on some data value --- ## Aesthetics: Setting vs. mapping Layers take arguments to control their appearance, such as point/line colors or transparency (`alpha` between 0 and 1). -- * Arguments like `color`, `size`, `linetype`, `shape`, `fill`, and `alpha` can be used directly on the layers (**setting aesthetics**), e.g. `geom_point(color = "red")`. See the [`ggplot2` documentation](http://docs.ggplot2.org/current/vignettes/ggplot2-specs.html) for options. These *don't depend on the data*. -- * Arguments inside `aes()` (**mapping aesthetics**) will *depend on the data*, e.g. `geom_point(aes(color = continent))`. -- * `aes()` in the `ggplot()` layer gives overall aesthetics to use in other layers, but can be changed on individual layers (including switching `x` or `y` to different variables) -- This may seem pedantic, but precise language makes searching for help easier. -- Now let's see all this jargon in action. --- ## Axis Labels, Points, No Background ### 1: Base Plot .pull-left[ .small[ ```r *ggplot(data = China, * aes(x = year, y = lifeExp)) ``` ] ] .pull-right[ <!-- --> ] .footnote[We initialize the plot with `ggplot()` and an aesthetic.] --- count: false ## Axis Labels, Points, No Background ### 2: Scatterplot .pull-left[ .small[ ```r ggplot(data = China, aes(x = year, y = lifeExp)) + * geom_point() ``` ] ] .pull-right[ <!-- --> ] .footnote[Add a scatterplot.] --- count: false ## Axis Labels, Points, No Background ### 3: Point Color and Size .pull-left[ .small[ ```r ggplot(data = China, aes(x = year, y = lifeExp)) + * geom_point(color = "red", size = 3) ``` ] ] .pull-right[ <!-- --> ] .footnote[Make the points large and red.] --- count: false ## Axis Labels, Points, No Background ### 4: X-Axis Label .pull-left[ .small[ ```r ggplot(data = China, aes(x = year, y = lifeExp)) + geom_point(color = "red", size = 3) + * xlab("Year") ``` ] ] .pull-right[ <!-- --> ] .footnote[Capitalize the x-axis label.] --- count: false ## Axis Labels, Points, No Background ### 5: Y-Axis Label .pull-left[ .small[ ```r ggplot(data = China, aes(x = year, y = lifeExp)) + geom_point(color = "red", size = 3) + xlab("Year") + * ylab("Life expectancy") ``` ] ] .pull-right[ <!-- --> ] .footnote[Clean up the y-axis label.] --- count: false ## Axis Labels, Points, No Background ### 6: Title .pull-left[ .small[ ```r ggplot(data = China, aes(x = year, y = lifeExp)) + geom_point(color = "red", size = 3) + xlab("Year") + ylab("Life expectancy") + * ggtitle("Life expectancy in China") ``` ] ] .pull-right[ <!-- --> ] .footnote[Add a title.] --- count: false ## Axis Labels, Points, No Background ### 7: Theme .pull-left[ .small[ ```r ggplot(data = China, aes(x = year, y = lifeExp)) + geom_point(color = "red", size = 3) + xlab("Year") + ylab("Life expectancy") + ggtitle("Life expectancy in China") + * theme_bw() ``` ] ] .pull-right[ <!-- --> ] .footnote[Pick a nicer theme.] --- count: false ## Axis Labels, Points, No Background ### 8: Text Size .pull-left[ .small[ ```r ggplot(data = China, aes(x = year, y = lifeExp)) + geom_point(color = "red", size = 3) + xlab("Year") + ylab("Life expectancy") + ggtitle("Life expectancy in China") + * theme_bw(base_size=18) ``` ] ] .pull-right[ <!-- --> ] .footnote[Increase the base text size.] --- # Plotting All Countries We have a plot we like for China... ... but what if we want *all the countries*? --- count: false # Plotting All Countries ### 1: A Mess! .pull-left[ .small[ ```r *ggplot(data = gapminder, aes(x = year, y = lifeExp)) + geom_point(color = "red", size = 3) + xlab("Year") + ylab("Life expectancy") + ggtitle("Life expectancy over time") + theme_bw(base_size=18) ``` ] ] .pull-right[ <!-- --> ] .footnote[We can't tell countries apart! Maybe we could follow *lines*?] --- count: false # Plotting All Countries ### 2: Lines .pull-left[ .small[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp)) + * geom_line(color = "red", size = 3) + xlab("Year") + ylab("Life expectancy") + ggtitle("Life expectancy over time") + theme_bw(base_size=18) ``` ] ] .pull-right[ <!-- --> ] .footnote[`ggplot2` doesn't know how to connect the lines!] --- count: false # Plotting All Countries ### 3: Grouping .pull-left[ .small[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, * group = country)) + geom_line(color = "red", size = 3) + xlab("Year") + ylab("Life expectancy") + ggtitle("Life expectancy over time") + theme_bw(base_size=18) ``` ] ] .pull-right[ <!-- --> ] .footnote[That looks more reasonable... but the lines are too thick!] --- count: false # Plotting All Countries ### 4: Size .pull-left[ .small[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) + * geom_line(color = "red") + xlab("Year") + ylab("Life expectancy") + ggtitle("Life expectancy over time") + theme_bw(base_size=18) ``` ] ] .pull-right[ <!-- --> ] .footnote[Much better... but maybe we can do highlight regional differences?] --- count: false # Plotting All Countries ### 5: Color .pull-left[ .small[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country, * color = continent)) + * geom_line() + xlab("Year") + ylab("Life expectancy") + ggtitle("Life expectancy over time") + theme_bw(base_size=18) ``` ] ] .pull-right[ <!-- --> ] .footnote[Patterns are obvious... but why not separate continents completely?] --- count: false # Plotting All Countries ### 6: Facets .pull-left[ .small[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country, color = continent)) + geom_line() + xlab("Year") + ylab("Life expectancy") + ggtitle("Life expectancy over time") + theme_bw(base_size=18) + * facet_wrap(~ continent) ``` ] ] .pull-right[ <!-- --> ] .footnote[Now the text is too big!] --- count: false # Plotting All Countries ### 7: Text Size .pull-left[ .small[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country, color = continent)) + geom_line() + xlab("Year") + ylab("Life expectancy") + ggtitle("Life expectancy over time") + * theme_bw() + facet_wrap(~ continent) ``` ] ] .pull-right[ <!-- --> ] .footnote[Looking good!] --- # Storing Plots We can assign a `ggplot` object to a name: ```r lifeExp_by_year <- ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country, color = continent)) + geom_line() + xlab("Year") + ylab("Life expectancy") + ggtitle("Life expectancy over time") + theme_bw() + facet_wrap(~ continent) ``` The graph won't be displayed when you do this. You can show the graph using a single line of code with just the object name, *or take the object and add more layers*. --- # Showing a Stored Graph ```r lifeExp_by_year ``` <!-- --> --- ## Adding a Layer ```r lifeExp_by_year + theme(legend.position = "bottom") ``` <!-- --> --- ## Common Problem: Overplotting Often we want a scatterplot of things that have discrete units. All those dots plot over each other! .small[ ```r ggplot(data = gapminder, aes(x = continent, y = year, color = continent)) + geom_point() ``` <!-- --> ] --- ## Fixing Overplotting with Jitter Inside `geom_point()`, `position = position_jitter(width=a, height=b)` shifts points up to `a` units horizontally and `b` units vertically. .small[ ```r ggplot(data = gapminder, aes(x = continent, y = year, color = continent)) + geom_point(position = position_jitter(width = 0.5, height = 2)) ``` <!-- --> ] --- # Histograms `ggplot2` uses `geom_histogram()` to produce histograms. This geometry will automatically bin data, but you can adjust the number of bins with `bins =`. ```r gapminder %>% ggplot(aes(x = lifeExp, fill = continent)) + geom_histogram(bins = 30) ``` <!-- --> --- # Changing the Axes We can modify the axes in a variety of ways, such as: * Change the `\(x\)` or `\(y\)` range using `xlim()` or `ylim()` layers * Change to a logarithmic or square-root scale on either axis: `scale_x_log10()`, `scale_y_sqrt()` * Change where the major/minor breaks are: `scale_x_continuous(breaks =, minor_breaks = )` --- # Axis Changes ```r ggplot(data = China, aes(x = year, y = gdpPercap)) + geom_line() + * scale_y_log10(breaks = c(1000, 2000, 3000, 4000, 5000), * labels = scales::dollar) + xlim(1940, 2010) + ggtitle("Chinese GDP per capita") ``` <!-- --> --- # Fonts Too Small? ```r ggplot(data = China, aes(x = year, y = lifeExp)) + geom_line() + ggtitle("Chinese life expectancy") + * theme_gray(base_size = 20) ``` <!-- --> --- # Text and Tick Adjustments Text size, labels, tick marks, etc. can be messed with more precisely using arguments to the `theme()` layer. Examples: * `plot.title = element_text(size = rel(2), hjust = 0)` makes the title twice as big as usual and left-aligns it * `axis.text.x = element_text(angle = 45)` rotates `\(x\)` axis labels * `axis.text = element_text(colour = "blue")` makes the `\(x\)` and `\(y\)` axis labels blue * `axis.ticks.length = unit(.5, "cm")` makes the axis ticks longer Note: `theme()` is a different layer than `theme_gray()` or `theme_bw()`, which you might also be using in a previous layer. See the [`ggplot2` documentation](http://docs.ggplot2.org/current/theme.html) for details. I recommend using `theme()` *after* `theme_bw()` or other *global themes*. --- # Scales for Color, Shape, etc. **Scales** are layers that control how the mapped aesthetics appear. You can modify these with a `scale_[aesthetic]_[option]()` layer where `[aesthetic]` is `color`, `shape`, `linetype`, `alpha`, `size`, `fill`, etc. and `[option]` is something like `manual`, `continuous` or `discrete` (depending on nature of the variable). Examples: * `scale_linetype_manual()`: manually specify the linetype for each different value * `scale_alpha_continuous()`: varies transparency over a continuous range * `scale_color_brewer(palette = "Spectral")`: uses a palette from <http://colorbrewer2.org> (great site for picking nice plot colors!) When confused... Google or StackOverflow it! --- ## Legend Name and Manual Colors .small[ ```r lifeExp_by_year + scale_color_manual( * name = "Which\ncontinent\nare we\nlooking at?", # \n adds a line break values = c("Africa" = "seagreen", "Americas" = "turquoise1", "Asia" = "royalblue", "Europe" = "violetred1", "Oceania" = "yellow")) ``` <!-- --> ] --- ## Fussy Manual Legend Example Code .small[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) + geom_line(alpha = 0.5, aes(color = "Country", size = "Country")) + * geom_line(stat = "smooth", method = "loess", aes(group = continent, color = "Continent", size = "Continent"), alpha = 0.5) + * facet_wrap(~ continent, nrow = 2) + * scale_color_manual(name = "Life Exp. for:", * values = c("Country" = "black", "Continent" = "blue")) + scale_size_manual(name = "Life Exp. for:", values = c("Country" = 0.25, "Continent" = 3)) + theme_minimal(base_size = 14) + ylab("Years") + xlab("") + ggtitle("Life Expectancy, 1952-2007", subtitle = "By continent and country") + * theme(legend.position=c(0.75, 0.2), axis.text.x = element_text(angle = 45)) ``` ] Wow, there's a lot going on here! * Two different `geom_line()` calls + One of them draws a [*loess* curve](https://en.wikipedia.org/wiki/Local_regression) * `facet_wrap()` to make a plot for each level of `continent` * Manual scales for size and color * Custom labels, titles, and rotated x axis text --- ## 1. Base Plot .smaller[ ```r *ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) # # # # # # # # # ``` <!-- --> ] --- count: false ## 2. Lines .smaller[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) + * geom_line() # # # # # # # # ``` <!-- --> ] --- count: false ## 3. Continent Average .smaller[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) + geom_line() + * geom_line(stat = "smooth", method = "loess", * aes(group = continent)) # # # # # # ``` <!-- --> ] --- count: false ## 4. Facets .smaller[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) + geom_line() + geom_line(stat = "smooth", method = "loess", aes(group = continent)) + * facet_wrap(~ continent, nrow = 2) # # # # # ``` <!-- --> ] --- count: false ## 5. Color Scale .smaller[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) + * geom_line(aes(color = "Country")) + geom_line(stat = "smooth", method = "loess", * aes(group = continent, color = "Continent")) + facet_wrap(~ continent, nrow = 2) + * scale_color_manual(name = "Life Exp. for:", values = c("Country" = "black", "Continent" = "blue")) # # # # ``` <!-- --> ] --- count: false ## 6. Size Scale .smaller[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) + * geom_line(aes(color = "Country", size = "Country")) + geom_line(stat = "smooth", method = "loess", * aes(group = continent, color = "Continent", size = "Continent")) + facet_wrap(~ continent, nrow = 2) + scale_color_manual(name = "Life Exp. for:", values = c("Country" = "black", "Continent" = "blue")) + * scale_size_manual(name = "Life Exp. for:", values = c("Country" = 0.25, "Continent" = 3)) # # # ``` <!-- --> ] --- count: false ## 7. Alpha (Transparency) .smaller[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) + * geom_line(alpha = 0.5, aes(color = "Country", size = "Country")) + geom_line(stat = "smooth", method = "loess", * aes(group = continent, color = "Continent", size = "Continent"), alpha = 0.5) + facet_wrap(~ continent, nrow = 2) + scale_color_manual(name = "Life Exp. for:", values = c("Country" = "black", "Continent" = "blue")) + scale_size_manual(name = "Life Exp. for:", values = c("Country" = 0.25, "Continent" = 3)) # # # ``` <!-- --> ] --- count: false ## 8. Theme and Labels .smaller[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) + geom_line(alpha = 0.5, aes(color = "Country", size = "Country")) + geom_line(stat = "smooth", method = "loess", aes(group = continent, color = "Continent", size = "Continent"), alpha = 0.5) + facet_wrap(~ continent, nrow = 2) + scale_color_manual(name = "Life Exp. for:", values = c("Country" = "black", "Continent" = "blue")) + scale_size_manual(name = "Life Exp. for:", values = c("Country" = 0.25, "Continent" = 3)) + * theme_minimal(base_size = 14) + ylab("Years") + xlab("") # # ``` <!-- --> ] --- count: false ## 9. Title and Subtitle .smaller[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) + geom_line(alpha = 0.5, aes(color = "Country", size = "Country")) + geom_line(stat = "smooth", method = "loess", aes(group = continent, color = "Continent", size = "Continent"), alpha = 0.5) + facet_wrap(~ continent, nrow = 2) + scale_color_manual(name = "Life Exp. for:", values = c("Country" = "black", "Continent" = "blue")) + scale_size_manual(name = "Life Exp. for:", values = c("Country" = 0.25, "Continent" = 3)) + theme_minimal(base_size = 14) + ylab("Years") + xlab("") + * ggtitle("Life Expectancy, 1952-2007", subtitle = "By continent and country") # ``` <!-- --> ] --- count: false ## 10. Angled Tick Values .smaller[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) + geom_line(alpha = 0.5, aes(color = "Country", size = "Country")) + geom_line(stat = "smooth", method = "loess", aes(group = continent, color = "Continent", size = "Continent"), alpha = 0.5) + facet_wrap(~ continent, nrow = 2) + scale_color_manual(name = "Life Exp. for:", values = c("Country" = "black", "Continent" = "blue")) + scale_size_manual(name = "Life Exp. for:", values = c("Country" = 0.25, "Continent" = 3)) + theme_minimal(base_size = 14) + ylab("Years") + xlab("") + ggtitle("Life Expectancy, 1952-2007", subtitle = "By continent and country") + * theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust = 1)) ``` <!-- --> ] .footnote[Note: Fewer values might be better than angled labels!] --- count: false ## 11. Legend Position .smaller[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, group = country)) + geom_line(alpha = 0.5, aes(color = "Country", size = "Country")) + geom_line(stat = "smooth", method = "loess", aes(group = continent, color = "Continent", size = "Continent"), alpha = 0.5) + facet_wrap(~ continent, nrow = 2) + scale_color_manual(name = "Life Exp. for:", values = c("Country" = "black", "Continent" = "blue")) + scale_size_manual(name = "Life Exp. for:", values = c("Country" = 0.25, "Continent" = 3)) + theme_minimal(base_size = 14) + ylab("Years") + xlab("") + ggtitle("Life Expectancy, 1952-2007", subtitle = "By continent and country") + * theme(legend.position=c(0.85, 0.1), axis.text.x = element_text(angle = 45, vjust = 1, hjust = 1)) ``` <!-- --> ] --- ## Fussy Manual Legend <!-- --> Observation: One could use `filter()` to identify the countries with dips in life expectancy and investigate. Know Your History: What happened in Africa in the early 1990s and Asia in the mid-1970s that might reduce life expectancy suddenly *for one country*? --- ## More on Customizing Legends You can move the legends around, flip their orientation, remove them altogether, etc. The [Cookbook for R website](http://www.cookbook-r.com/Graphs/Legends_%28ggplot2%29) is a good resource for questions such as changing legend labels. --- # Saving `ggplot` Plots When you knit an R Markdown file, any plots you make are automatically saved in the "figure" folder in `.png` format. If you want to save another copy (perhaps of a different file type for use in a manuscript), use `ggsave()`: ```r ggsave("I_saved_a_file.pdf", plot = lifeExp_by_year, height = 3, width = 5, units = "in") ``` If you didn't manually set font sizes, these will usually come out at a reasonable size given the dimensions of your output file. **Bad/non-reproducible way**<sup>1</sup>: choose *Export* on the plot preview or take a screenshot / snip. .footnote[[1] I still do this for quick emails of simple plots. Bad me!] --- class: inverse # Plotting Model Results --- # `geom_smooth()` I have used `geom_smooth()` in multiple prior examples. `geom_smooth()` generates "smoothed conditional means" including loess curves and generalized additive models (GAMs). -- Note, however, that most regression models are conditional mean models, such as ordinary least squares, generalized linear models. -- We can use `geom_smooth()` to add a layer depicting common bivariate models. --- # Default `geom_smooth()` ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent)) + geom_point(position = position_jitter(1,0), size = 0.5) + geom_smooth() ``` <!-- --> By default, `geom_smooth()` chooses either a loess smoother (N < 1000) or a GAM depending on the number of observations. --- # Linear `glm` ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent)) + geom_point(position = position_jitter(1,0), size = 0.5) + * geom_smooth(method = "glm", formula = y ~ x) ``` <!-- --> We could also fit a standard linear model using either `method = "glm"` or `method = "lm"` and a formula like `y ~ x`. --- # Polynomial `glm` ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent)) + geom_point(position = position_jitter(1,0), size = 0.5) + * geom_smooth(method = "glm", formula = y ~ poly(x, 2)) ``` <!-- --> `poly(x, 2)` produces a quadratic model which contains a linear term (`x`) and a quadratic term (`x^2`). --- # `ggeffects` If we want to look at more complex models, we can use `ggeffects` to create and plot tidy *marginal effects*. That is, tidy dataframes of *ranges* of predicted values that can be fed straight into `ggplot2` for plotting model results. We will focus on two `ggeffects` functions: * `ggpredict()` - Computes predicted values for the outcome variable at margins of specific variables. * `plot.ggeffects()` - A plot method for `ggeffects` objects (like `ggredict()` output) ```r library(ggeffects) ``` --- # Quick Simulated Data To show off `ggeffects`, I need a data frame with a variety of numeric and categorical variables with strong relationships. It is easiest to just simulate it: ```r ex_dat <- data.frame(num1 = rnorm(200, 1, 2), fac1 = sample(c(1, 2, 3), 200, TRUE), num2 = rnorm(200, 0, 3), fac2 = sample(c(1, 2))) %>% mutate(yn = num1 * 0.5 + fac1 * 1.1 + num2 * 0.7 + fac2 - 1.5 + rnorm(200, 0, 2)) %>% mutate(yb = as.numeric(yn > mean(yn))) %>% mutate(fac1 = factor(fac1, labels = c("A", "B", "C")), fac2 = factor(fac2, labels = c("Yes", "No"))) ``` --- # `ggpredict()` When you run `ggpredict()`, it produces a dataframe with a row for every unique value of a supplied predictor ("independent") variable (`term`). Each row contains an expected (estimated) value for the outcome ("dependent") variable, plus confidence intervals. ```r lm_1 <- lm(yn ~ num1 + fac1, data = ex_dat) lm_1_est <- ggpredict(lm_1, terms = "num1") ``` If desired, the argument `interval="prediction"` will give predicted intervals instead. --- #`ggpredict()` output .smallish[ ```r lm_1_est ``` ``` ## ## # Predicted values of yn ## # x = num1 ## ## x predicted std.error conf.low conf.high ## -6 -2.147 0.782 -3.678 -0.615 ## -4 -1.180 0.606 -2.369 0.008 ## -2 -0.214 0.450 -1.096 0.667 ## 0 0.752 0.339 0.088 1.416 ## 2 1.718 0.324 1.083 2.353 ## 4 2.685 0.416 1.869 3.500 ## 6 3.651 0.565 2.544 4.758 ## 8 4.617 0.736 3.174 6.061 ## ## Adjusted for: ## * fac1 = A ``` ] --- # `plot()` for `ggpredict()` `ggeffects` features a `plot()` *method*, `plot.ggeffects()`, which produces a ggplot when you give `plot()` output from `ggpredict()`. .small[ ```r plot(lm_1_est) ``` <!-- --> ] --- # Grouping with `ggpredict()` When using a vector of `terms`, `ggeffects` will plot the first along the x-axis and use others for *grouping*. Note we can pipe a model into `ggpredict()`! .small[ ```r glm(yb ~ num1 + fac1 + num2 + fac2, data = ex_dat, family=binomial(link = "logit")) %>% ggpredict(terms = c("num1", "fac1")) %>% plot() ``` <!-- --> ] --- # Faceting with `ggpredict()` You can add `facet=TRUE` to the `plot()` call to facet over *grouping terms*. .small[ ```r glm(yb ~ num1 + fac1 + num2 + fac2, data = ex_dat, family = binomial(link = "logit")) %>% ggpredict(terms = c("num1", "fac1")) %>% plot(facet=TRUE) ``` <!-- --> ] --- # Counterfactual Values You can add values in square brackets in the `terms=` argument to specify counterfactual values. .small[ ```r glm(yb ~ num1 + fac1 + num2 + fac2, data=ex_dat, family=binomial(link="logit")) %>% ggpredict(terms = c("num1 [-1,0,1]", "fac1 [A,B]")) %>% plot(facet=TRUE) ``` <!-- --> ] --- # Representative Values You can also use `[meansd]` or `[minmax]` to set representative values. .small[ ```r glm(yb ~ num1 + fac1 + num2 + fac2, data = ex_dat, family = binomial(link = "logit")) %>% ggpredict(terms = c("num1 [meansd]", "num2 [minmax]")) %>% plot(facet=TRUE) ``` <!-- --> ] --- # Dot plots with `ggpredict()` `ggpredict` will produce dot plots with error bars for categorical predictors. .small[ ```r lm(yn ~ fac1 + fac2, data = ex_dat) %>% ggpredict(terms=c("fac1", "fac2")) %>% plot() ``` <!-- --> ] --- # Notes on `ggeffects` There is a lot more to the `ggeffects` package that you can see in [the package vignette](https://cran.r-project.org/web/packages/ggeffects/vignettes/marginaleffects.html) and the [github repository](https://github.com/strengejacke/ggeffects). This includes, but is not limited to: * Predicted values for polynomial and interaction terms * Getting predictions from models from dozens of other packages * Sending `ggeffects` objects to `ggplot2` to freely modify plots --- # Bonus Plot `ggplot2` is well suited to making complex, publication ready plots. This is the complete syntax for one plot from a recent article of mine.<sup>1</sup> .small[ ```r ggplot(estimated_pes, aes(x = Target, y = PE, group = Reporter)) + facet_grid(`Crime Type` ~ Neighborhood) + geom_errorbar(aes(ymin = LB, ymax = UB), position = position_dodge(width = .4), size = 0.75, width = 0.15) + geom_point(shape = 21, position = position_dodge(width = .4), size = 2, aes(fill = Reporter)) + scale_fill_manual("Reporter", values = c("Any White" = "white", "All Black" = "black")) + ggtitle("Figure 3. Probability of Arrest", subtitle = "by Reporter and Target Race, Neighborhood and Crime Type") + xlab("Race of Target") + ylab("Estimated Probability") + theme_bw() + theme(legend.position = c(0.86, 0.15), legend.background = element_rect(color = 1)) ``` ] .footnote[ [1] [Lanfear, Charles C., Lindsey R. Beach, Timothy A. Thomas. 2018. "Formal Social Control in Changing Neighborhoods: Racial Implications of Neighborhood Context on Reactive Policing." *City & Community* 17(4):1075-1099](https://onlinelibrary.wiley.com/doi/10.1111/cico.12346) ] --- .center[ <img src="img/arrest_pe_plot.png" style="width: 80%;"/> ] --- class: inverse # Book Recommendation .pull-left[  ] .pull-right[ * Targeted at Social Scientists without technical backgrounds * Teaches good visualization principles * Uses R, `ggplot2`, and `tidyverse` * [Free online version!](https://socviz.co/) * Affordable in print ] --- # Questions