class: center, top, title-slide # CSSS508, Week 10 ## Model Results and Reproducibility ### Chuck Lanfear ### Jun 2, 2021<br>Updated: Jun 1, 2021 --- # Topics for Today ### Working with Model Results * Tidy model output with `broom` * Visualizing models with `ggeffects` * Tables with `gt`, `modelsummary`, and `gtsummary` ### Reproducible Research ### Best Practices * Organization * Portability * Version Control ### Wrapping up the course --- class: inverse # Working with Model Results --- # `broom` `broom` is a package that "tidies up" the output from models such a `lm()` and `glm()`. It has a small number of key functions: * `tidy()` - Creates a dataframe summary of a model. * `augment()` - Adds columns—such as fitted values—to the data used in the model. * `glance()` - Provides one row of fit statistics for models. ```r library(broom) ``` --- # Model Output is a List `lm()` and `summary()` produce lists as output, which cannot go directly into tidyverse functions, particularly those in `ggplot2`. .smaller[ ```r lm_1 <- lm(yn ~ num1 + fac1, data = ex_dat) summary(lm_1) ``` ``` ## ## Call: ## lm(formula = yn ~ num1 + fac1, data = ex_dat) ## ## Residuals: ## Min 1Q Median 3Q Max ## -9.9971 -1.7452 -0.1423 2.1099 7.4219 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 1.1632 0.3826 3.041 0.00268 ** ## num1 0.6932 0.1044 6.643 2.96e-10 *** ## fac1B 0.7495 0.5177 1.448 0.14932 ## fac1C 2.2360 0.5009 4.464 1.36e-05 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 2.945 on 196 degrees of freedom ## Multiple R-squared: 0.2364, Adjusted R-squared: 0.2247 ## F-statistic: 20.22 on 3 and 196 DF, p-value: 1.849e-11 .... ``` ] --- # Model Output Varies! Each type of model also produces somewhat different output, so you can't just reuse the same code to handle output from every model. .smaller[ ```r glm_1 <- glm(yb ~ num1 + fac1, data = ex_dat, family=binomial(link="logit")) summary(glm_1) ``` ``` ## ## Call: ## glm(formula = yb ~ num1 + fac1, family = binomial(link = "logit"), ## data = ex_dat) ## ## Deviance Residuals: ## Min 1Q Median 3Q Max ## -1.994 -1.056 -0.421 1.020 2.207 ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -1.09058 0.30014 -3.634 0.000280 *** ## num1 0.38134 0.08786 4.340 1.42e-05 *** ## fac1B 0.49142 0.37958 1.295 0.195450 ## fac1C 1.29566 0.37993 3.410 0.000649 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for binomial family taken to be 1) ## .... ``` ] --- # `broom::tidy()` `tidy()` produces similar output, but as a dataframe. ```r lm_1 %>% tidy() ``` ``` ## # A tibble: 4 x 5 ## term estimate std.error statistic p.value ## <chr> <dbl> <dbl> <dbl> <dbl> ## 1 (Intercept) 1.16 0.383 3.04 2.68e- 3 ## 2 num1 0.693 0.104 6.64 2.96e-10 ## 3 fac1B 0.749 0.518 1.45 1.49e- 1 ## 4 fac1C 2.24 0.501 4.46 1.36e- 5 ``` Each type of model (e.g. `glm`, `lmer`) has a different *method* with its own additional arguments. See `?tidy.lm` for an example. --- # `broom::tidy()` This output is also completely identical between different models. This can be very useful and important if running models with different test statistics... or just running a lot of models! ```r glm_1 %>% tidy() ``` ``` ## # A tibble: 4 x 5 ## term estimate std.error statistic p.value ## <chr> <dbl> <dbl> <dbl> <dbl> ## 1 (Intercept) -1.09 0.300 -3.63 0.000280 ## 2 num1 0.381 0.0879 4.34 0.0000142 ## 3 fac1B 0.491 0.380 1.29 0.195 ## 4 fac1C 1.30 0.380 3.41 0.000649 ``` --- # `broom::glance()` `glance()` produces dataframes of fit statistics for models. If you run many models, you can compare each model row-by-row in each column... or even plot their different fit statistics to allow holistic comparison. .small[ ```r glance(lm_1) ``` ``` ## # A tibble: 1 x 12 ## r.squared adj.r.squared sigma statistic p.value df logLik AIC ## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 0.236 0.225 2.95 20.2 1.85e-11 3 -498. 1006. ## # ... with 4 more variables: BIC <dbl>, deviance <dbl>, ## # df.residual <int>, nobs <int> ``` ] --- # `broom::augment()` `augment()` takes values generated by a model and adds them back to the original data. This includes fitted values, residuals, and leverage statistics. .small[ ```r augment(lm_1) %>% head() ``` ``` ## # A tibble: 6 x 9 ## yn num1 fac1 .fitted .resid .hat .sigma .cooksd .std.resid ## <dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 -1.10 2.26 B 3.48 -4.58 0.0176 2.93 0.0111 -1.57 ## 2 4.62 -2.52 C 1.65 2.97 0.0301 2.94 0.00815 1.02 ## 3 -2.40 0.101 B 1.98 -4.39 0.0181 2.94 0.0104 -1.50 ## 4 3.42 3.01 B 4.00 -0.578 0.0202 2.95 0.000203 -0.198 ## 5 8.07 0.512 C 3.75 4.32 0.0146 2.94 0.00810 1.48 ## 6 5.50 2.12 C 4.87 0.634 0.0158 2.95 0.000189 0.217 ``` ] See `?augment.lm` for examples of what `augment()` can do. --- # The Power of `broom` The real advantage of `broom` becomes apparent when running many models at once. Here we run separate models for each level of `fac1`: .smallish[ ```r ex_dat %>% * nest_by(fac1) %>% mutate(model = list(lm(yn ~ num1 + fac2, data = data))) %>% summarize(tidy(model), .groups = "drop") ``` ``` ## # A tibble: 9 x 6 ## fac1 term estimate std.error statistic p.value ## <fct> <chr> <dbl> <dbl> <dbl> <dbl> ## 1 A (Intercept) 0.00555 0.636 0.00873 0.993 ## 2 A num1 0.797 0.215 3.71 0.000433 ## 3 A fac2No 2.26 0.788 2.87 0.00558 ## 4 B (Intercept) 1.39 0.587 2.37 0.0211 ## 5 B num1 0.713 0.171 4.17 0.000103 ## 6 B fac2No 0.945 0.751 1.26 0.213 ## 7 C (Intercept) 3.04 0.464 6.55 0.00000000968 ## 8 C num1 0.693 0.153 4.52 0.0000257 ## 9 C fac2No 0.690 0.606 1.14 0.259 ``` ] .footnote[`nest_by()` nests data into a list column by levels of `fac1`.] --- class: inverse # Plotting Model Results --- # `geom_smooth()` I have used `geom_smooth()` in many past examples. `geom_smooth()` generates "smoothed conditional means" including loess curves and generalized additive models (GAMs). -- Note, however, that most regression models are conditional mean models, such as ordinary least squares and generalized linear models. -- We can use `geom_smooth()` to add a layer depicting common bivariate models. We'll look at this with the `gapminder` data from Week 2. ```r library(gapminder) ``` --- # Default `geom_smooth()` ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent)) + geom_point(position = position_jitter(1,0), size = 0.5) + geom_smooth() ``` <!-- --> By default, `geom_smooth()` chooses either a loess smoother (N < 1000) or a GAM depending on the number of observations. --- # Linear `glm` ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent)) + geom_point(position = position_jitter(1,0), size = 0.5) + * geom_smooth(method = "glm", formula = y ~ x) ``` <!-- --> We could also fit a standard linear model using either `method = "glm"` or `method = "lm"` and a formula like `y ~ x`. --- # Polynomial `glm` ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent)) + geom_point(position = position_jitter(1,0), size = 0.5) + * geom_smooth(method = "glm", formula = y ~ poly(x, 2)) ``` <!-- --> `poly(x, 2)` produces a quadratic model which contains a linear term (`x`) and a quadratic term (`x^2`). --- # More Complex Models What if we want something more complex than a bivariate model? What if we have a statistically complex model, like nonlinear probability model or multilevel model? We need to go beyond `geom_smooth()`! --- # But first, vocab! We are often interested in what might happen if some variables take particular values, often ones not seen in the actual data. -- When we set variables to certain values, we refer to them as **counterfactual values** or just **counterfactuals**. -- For example, if we know nothing about a new observation, our prediction for that estimate is often based on assuming every variable is at its mean. -- Sometimes, however, we might have very specific questions which require setting (possibly many) combinations of variables to particular values and making an estimate or prediction. -- Providing specific estimates, conditional on values of covariates, is a nice way to summarize results, particularly for models with unintuitive parameters (e.g. logit models). --- class: inverse # `ggeffects` --- # `ggeffects` If we want to look at more complex models, we can use `ggeffects` to create and plot tidy *marginal effects*. That is, tidy dataframes of *ranges* of predicted values that can be fed straight into `ggplot2` for plotting model results. We will focus on two `ggeffects` functions: * `ggpredict()` - Computes predicted values for the outcome variable at margins of specific variables. * `plot.ggeffects()` - A plot method for `ggeffects` objects (like `ggredict()` output) ```r library(ggeffects) ``` --- # Quick Simulated Data To best show off `ggeffects`, I need a data frame with numeric and categorical variables with strong relationships. It is easiest to just simulate it: ```r ex_dat <- data.frame(num1 = rnorm(200, 1, 2), fac1 = sample(c(1, 2, 3), 200, TRUE), num2 = rnorm(200, 0, 3), fac2 = sample(c(1, 2))) %>% mutate(yn = num1 * 0.5 + fac1 * 1.1 + num2 * 0.7 + fac2 - 1.5 + rnorm(200, 0, 2)) %>% mutate(yb = as.numeric(yn > mean(yn))) %>% mutate(fac1 = factor(fac1, labels = c("A", "B", "C")), fac2 = factor(fac2, labels = c("Yes", "No"))) ``` Now we can get `ggpredict`ing! --- # `ggpredict()` When you run `ggpredict()`, it produces a dataframe with a row for every unique value of a supplied predictor ("independent") variable (`term`). Each row contains an expected (estimated) value for the outcome ("dependent") variable, plus confidence intervals. ```r lm_1 <- lm(yn ~ num1 + fac1, data = ex_dat) lm_1_est <- ggpredict(lm_1, terms = "num1") ``` If desired, the argument `interval="prediction"` will give predicted intervals instead. --- #`ggpredict()` output .smallish[ ```r lm_1_est ``` ``` ## # Predicted values of yn ## # x = num1 ## ## x | Predicted | 95% CI ## ------------------------------ ## -6 | -0.68 | [-2.23, 0.87] ## -4 | 0.03 | [-1.16, 1.22] ## -2 | 0.73 | [-0.15, 1.61] ## 0 | 1.43 | [ 0.75, 2.12] ## 2 | 2.14 | [ 1.41, 2.87] ## 4 | 2.84 | [ 1.87, 3.81] ## 6 | 3.55 | [ 2.24, 4.85] ## 8 | 4.25 | [ 2.58, 5.93] ## ## Adjusted for: ## * fac1 = A ``` ] --- # `plot()` for `ggpredict()` `ggeffects` features a `plot()` *method*, `plot.ggeffects()`, which produces a ggplot when you give `plot()` output from `ggpredict()`. .small[ ```r plot(lm_1_est) ``` <!-- --> ] --- # Grouping with `ggpredict()` When using a vector of `terms`, `ggeffects` will plot the first along the x-axis and use others for *grouping*. Note we can pipe a model into `ggpredict()`! .small[ ```r glm(yb ~ num1 + fac1 + num2 + fac2, data = ex_dat, family=binomial(link = "logit")) %>% ggpredict(terms = c("num1", "fac1")) %>% plot() ``` <!-- --> ] --- # Faceting with `ggpredict()` You can add `facet=TRUE` to the `plot()` call to facet over *grouping terms*. .small[ ```r glm(yb ~ num1 + fac1 + num2 + fac2, data = ex_dat, family = binomial(link = "logit")) %>% ggpredict(terms = c("num1", "fac1")) %>% plot(facet=TRUE) ``` <!-- --> ] --- # Counterfactual Values You can add values in square brackets in the `terms=` argument to specify counterfactual values. .small[ ```r glm(yb ~ num1 + fac1 + num2 + fac2, data=ex_dat, family=binomial(link="logit")) %>% ggpredict(terms = c("num1 [-1,0,1]", "fac1 [A,B]")) %>% plot(facet=TRUE) ``` <!-- --> ] --- # Representative Values You can also use `[meansd]` or `[minmax]` to set representative values. .small[ ```r glm(yb ~ num1 + fac1 + num2 + fac2, data = ex_dat, family = binomial(link = "logit")) %>% ggpredict(terms = c("num1 [meansd]", "num2 [minmax]")) %>% plot(facet=TRUE) ``` <!-- --> ] --- # Dot plots with `ggpredict()` `ggpredict` will produce dot plots with error bars for categorical predictors. .small[ ```r lm(yn ~ fac1 + fac2, data = ex_dat) %>% ggpredict(terms=c("fac1", "fac2")) %>% plot() ``` <!-- --> ] --- # Notes on `ggeffects` There is a lot more to the `ggeffects` package that you can see in [the package vignette](https://cran.r-project.org/web/packages/ggeffects/vignettes/marginaleffects.html) and the [github repository](https://github.com/strengejacke/ggeffects). This includes, but is not limited to: * Predicted values for polynomial and interaction terms * Getting predictions from models from dozens of other packages * Sending `ggeffects` objects to `ggplot2` to freely modify plots If you need to do something more complex then `ggeffects` allows, see the [Advanced Counterfactuals](http://clanfear.github.io/CSSS508/Lectures/Week10/CSSS508_Advanced_Counterfactuals.html) slides here.<sup>1</sup> .footnote[[1] This is a bit out-of-date but the approach works and will give you an idea. Some day I'll make it into a package!] --- class: inverse # Making Tables --- # `pander` Regression Tables We've used `pander` to create nice tables for dataframes. But `pander` has *methods* to handle all sort of objects that you might want displayed nicely. This includes model output, such as from `lm()`, `glm()`, and `summary()`. ```r library(pander) ``` --- # `pander()` and `lm()` You can send an `lm()` object straight to `pander`: ```r pander(lm_1) ``` | | Estimate | Std. Error | t value | Pr(>t) | |:----------------|:--------:|:----------:|:-------:|:---------:| | **(Intercept)** | 37.23 | 1.599 | 23.28 | 2.565e-20 | | **wt** | -3.878 | 0.6327 | -6.129 | 1.12e-06 | | **hp** | -0.03177 | 0.00903 | -3.519 | 0.001451 | Table: Fitting linear model: mpg ~ wt + hp --- # `pander()` and `summary()` You can do this with `summary()` as well, for added information: ```r pander(summary(lm_1)) ``` | | Estimate | Std. Error | t value | Pr(>t) | |:----------------|:--------:|:----------:|:-------:|:---------:| | **(Intercept)** | 37.23 | 1.599 | 23.28 | 2.565e-20 | | **wt** | -3.878 | 0.6327 | -6.129 | 1.12e-06 | | **hp** | -0.03177 | 0.00903 | -3.519 | 0.001451 | | Observations | Residual Std. Error | `\(R^2\)` | Adjusted `\(R^2\)` | |:------------:|:-------------------:|:------:|:--------------:| | 32 | 2.593 | 0.8268 | 0.8148 | Table: Fitting linear model: mpg ~ wt + hp --- # Advanced Tables `pander` tables are great for basic `rmarkdown` documents, but they're not generally publication ready. We're going to talk about a few different approaches for making nicer tables: * `gt` from RStudio for general table construction * `modelsummary` for creating model tables * `gtsummary` for creating data summaries --- # `gt` If you need to more customizability or different output types, [RStudio's `gt` package](https://gt.rstudio.com/) is a new and powerful system for creating tables from dataframes. We'll use `dplyr`'s built-in `starwars` data for some examples. .smallish[ ```r library(gt) tes_chars <- starwars %>% unnest(films) %>% unnest(starships, keep_empty=TRUE) %>% filter(films == "The Empire Strikes Back") %>% select(name, species, starships, mass, height) %>% distinct(name, .keep_all = TRUE) %>% mutate(starships = ifelse(name == "Obi-Wan Kenobi" | is.na(starships), "No Ship", starships)) glimpse(tes_chars) ``` ``` ## Rows: 16 ## Columns: 5 ## $ name <chr> "Luke Skywalker", "C-3PO", "R2-D2", "Darth Vader",~ ## $ species <chr> "Human", "Droid", "Droid", "Human", "Human", "Huma~ ## $ starships <chr> "X-wing", "No Ship", "No Ship", "TIE Advanced x1",~ ## $ mass <dbl> 77.0, 75.0, 32.0, 136.0, 49.0, 77.0, 112.0, 80.0, ~ ## $ height <int> 172, 167, 96, 202, 150, 182, 228, 180, 170, 66, 17~ ``` ] --- .pull-left[ ## Initialize .small[ ```r tes_chars %>% gt() ``` ] `gt()` just renders the dataframe as a table using markdown. ] .smallest[ .pull-right[ .image-full[ 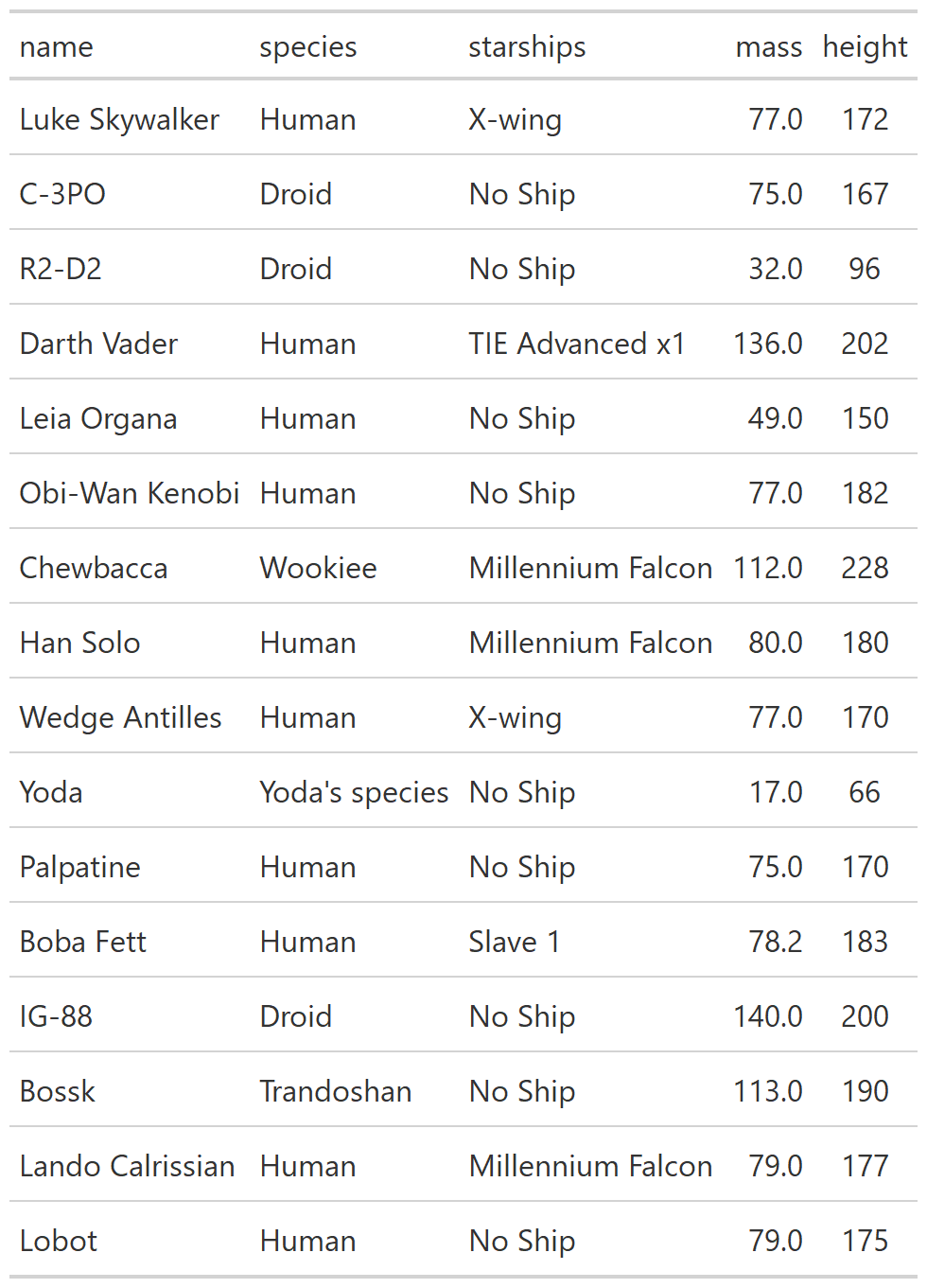<!-- --> ] ] ] --- count: false .pull-left[ ## 2: Grouping .small[ ```r tes_chars %>% group_by(starships) %>% gt() ``` ] If we `group_by()` first, the levels of the grouping variable become grouping rows in the table. ] .smallest[ .pull-right[ .image-tall[ 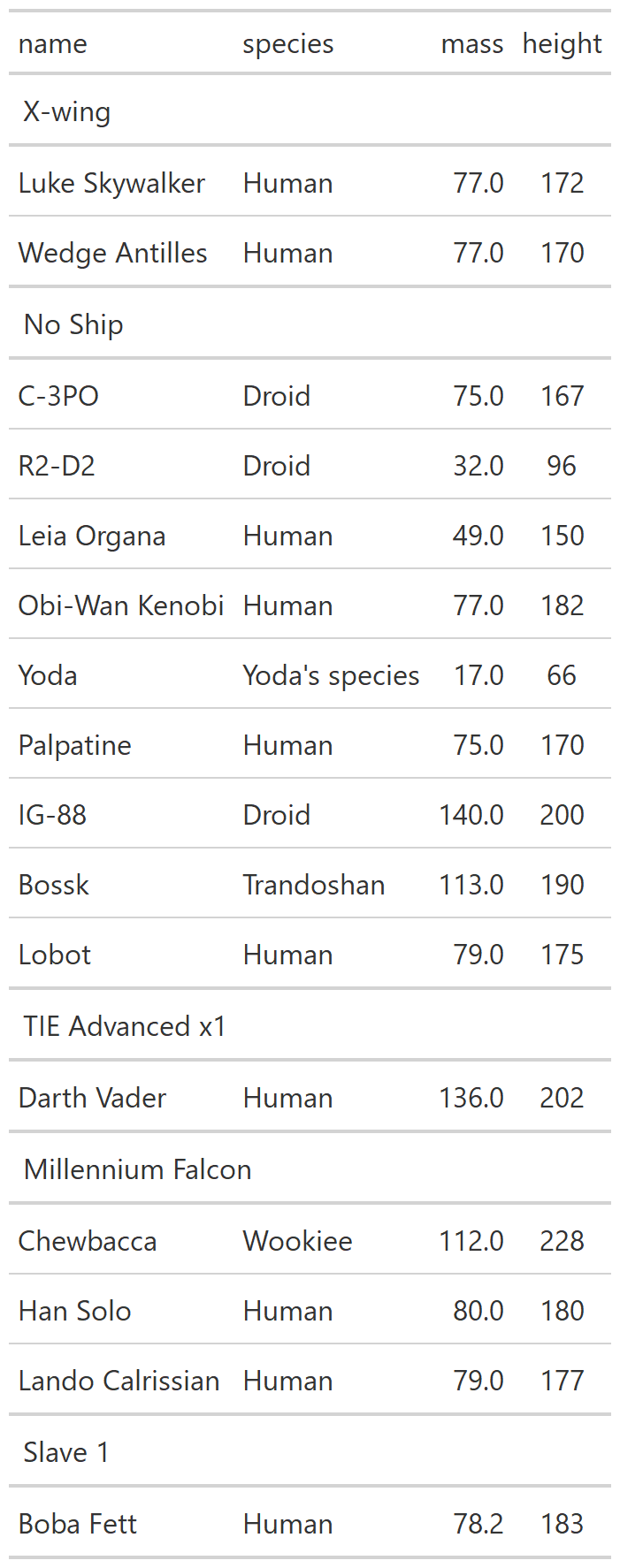<!-- --> ] ] ] --- count: false .pull-left[ ## 3: Rownames .small[ ```r tes_chars %>% group_by(starships) %>% gt(rowname_col = "name") ``` ] Designating a `rowname_col` removes that column's name and divides the values from the measures to the right. ] .smallest[ .pull-right[ .image-tall[ 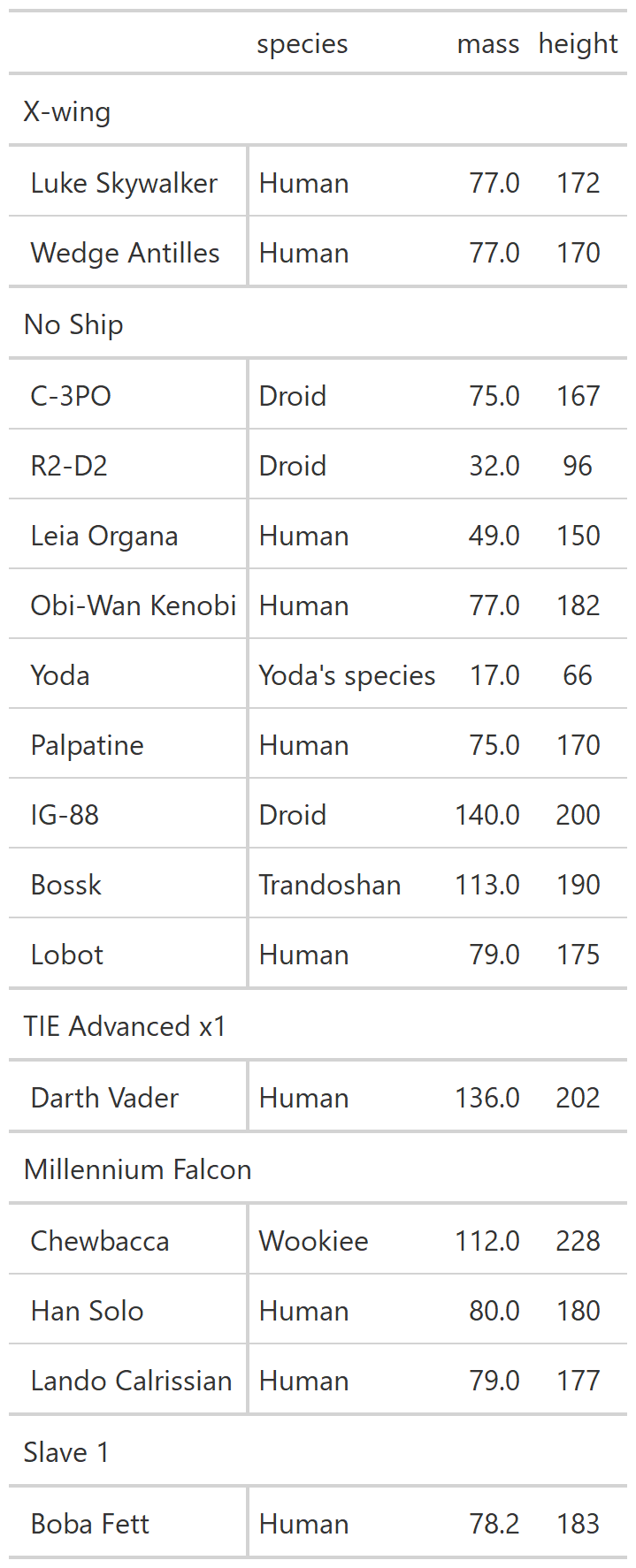<!-- --> ] ] ] --- count: false .pull-left[ ## Header .small[ ```r tes_chars %>% group_by(starships) %>% gt(rowname_col = "name") %>% tab_header( title = "Star Wars Characters", subtitle = "The Empire Strikes Back" ) ``` ] `tab_header()` can add titles and subtitles. ] .smallest[ .pull-right[ .image-tall[ 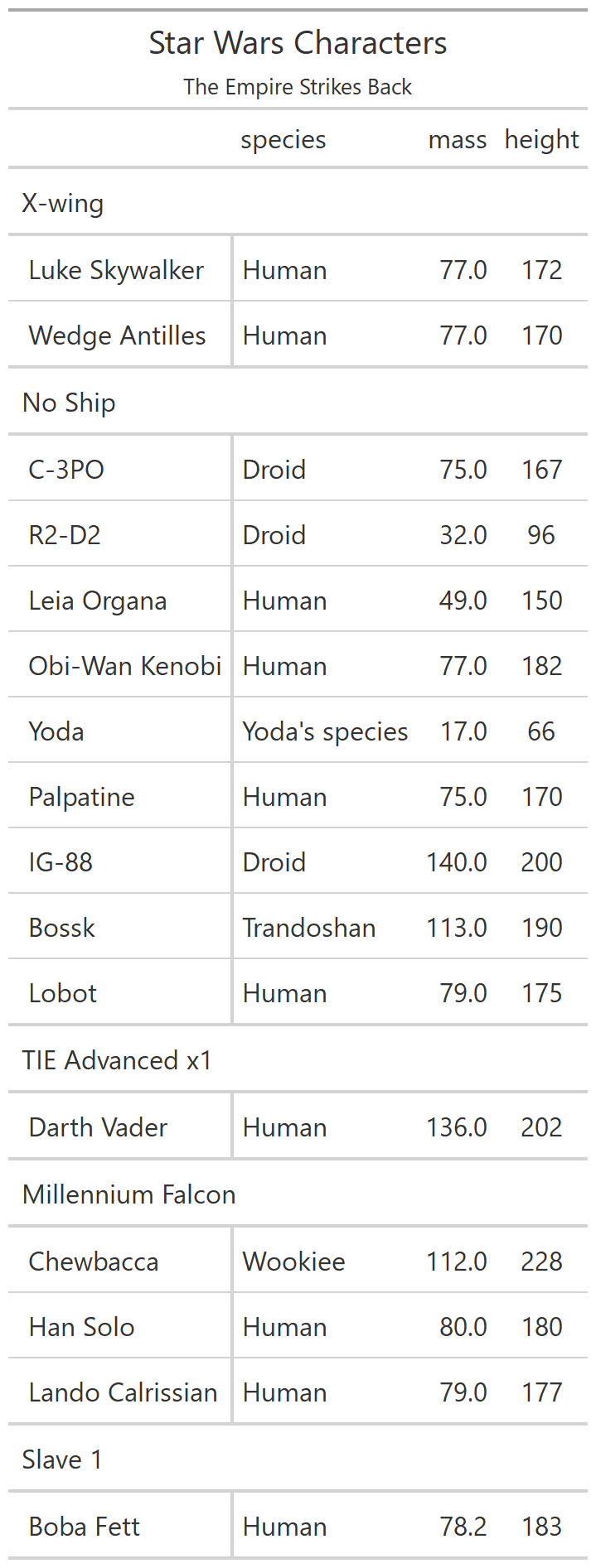<!-- --> ] ] ] --- count: false .pull-left[ ## Spanner .small[ ```r tes_chars %>% group_by(starships) %>% gt(rowname_col = "name") %>% tab_header( title = "Star Wars Characters", subtitle = "The Empire Strikes Back" ) %>% tab_spanner( label = "Vitals", columns = vars(mass, height) ) ``` ] A `tab_spanner()` lets us group columns together. ] .smallest[ .pull-right[ .image-tall[ 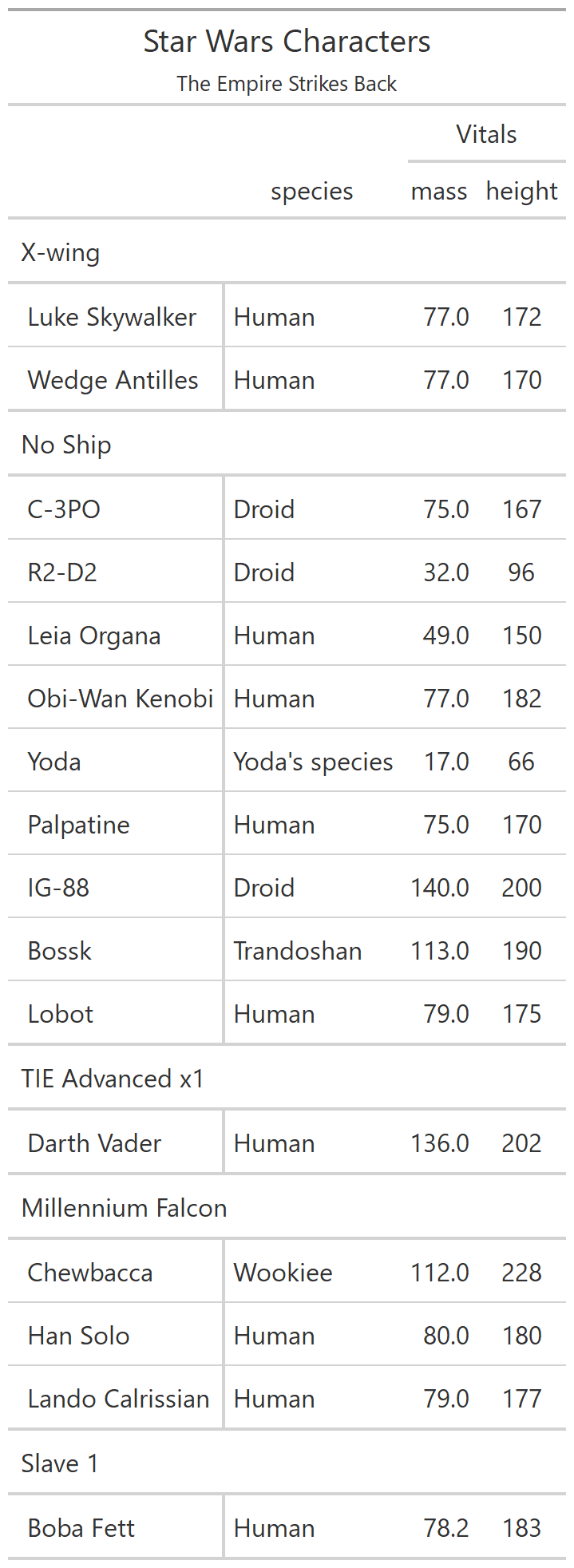<!-- --> ] ] ] --- count: false .pull-left[ ## Column Labels .small[ ```r tes_chars %>% group_by(starships) %>% gt(rowname_col = "name") %>% tab_header( title = "Star Wars Characters", subtitle = "The Empire Strikes Back" ) %>% tab_spanner( label = "Vitals", columns = vars(mass, height) ) %>% cols_label( mass = "Mass (kg)", height = "Height (cm)", species = "Species" ) ``` ] We can directly change column names with `cols_label()` ] .smallest[ .pull-right[ .image-tall[ 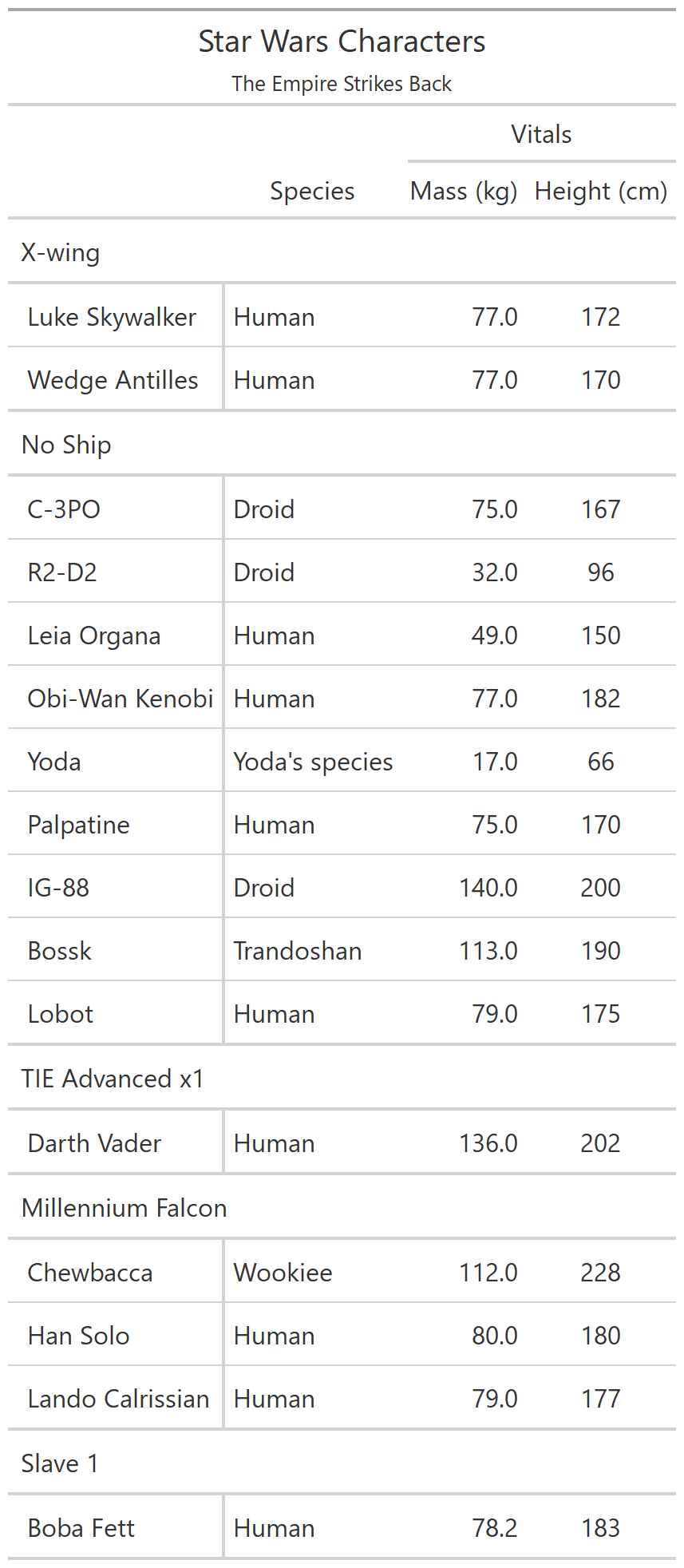<!-- --> ] ] ] --- count: false .pull-left[ ## Number Format .small[ ```r tes_chars %>% group_by(starships) %>% gt(rowname_col = "name") %>% tab_header( title = "Star Wars Characters", subtitle = "The Empire Strikes Back" ) %>% tab_spanner( label = "Vitals", columns = vars(mass, height) ) %>% cols_label( mass = "Mass (kg)", height = "Height (cm)", species = "Species" ) %>% fmt_number( columns = vars(mass), decimals = 0) ``` ] We can adjust cell formats too. ] .smallest[ .pull-right[ .image-tall[ 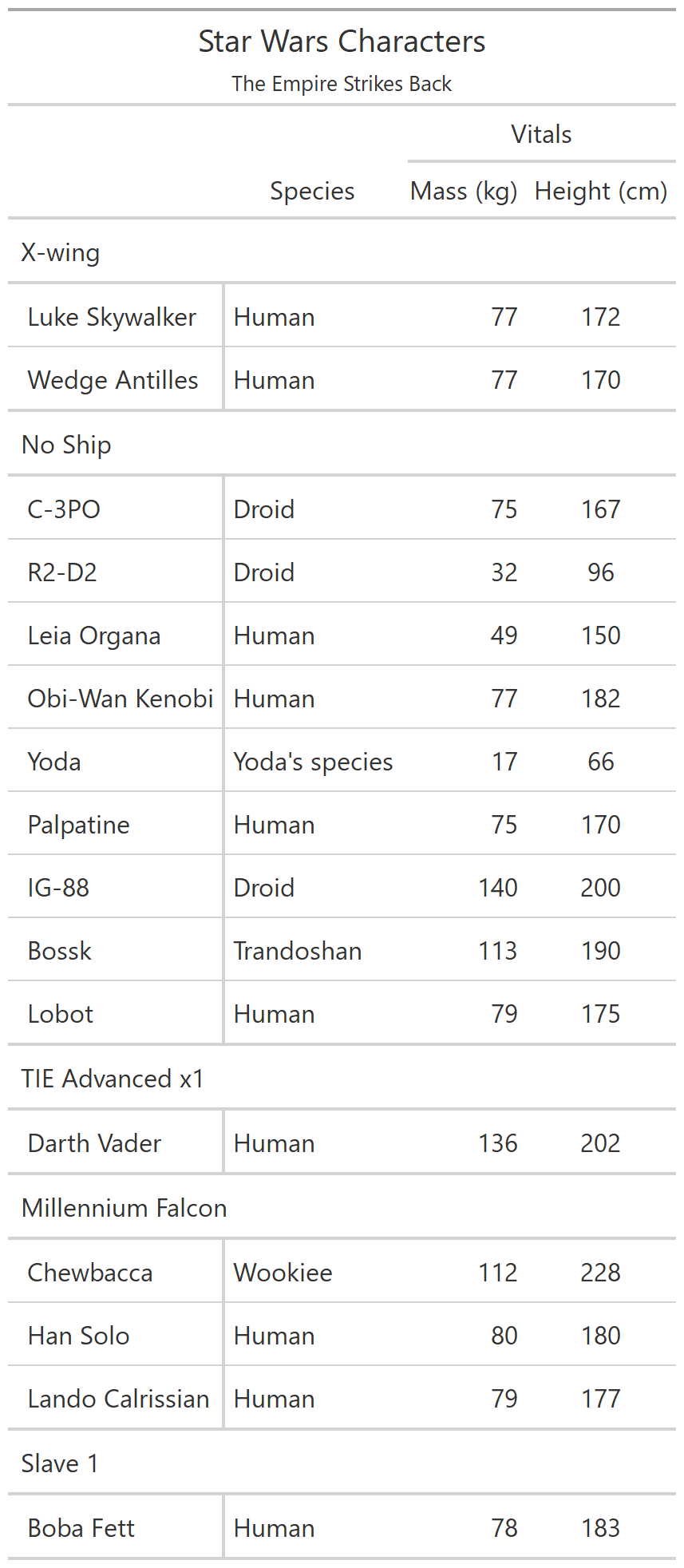<!-- --> ] ] ] --- count: false .pull-left[ ## Align Columns .small[ ```r tes_chars %>% group_by(starships) %>% gt(rowname_col = "name") %>% tab_header( title = "Star Wars Characters", subtitle = "The Empire Strikes Back" ) %>% tab_spanner( label = "Vitals", columns = vars(mass, height) ) %>% cols_label( mass = "Mass (kg)", height = "Height (cm)", species = "Species" ) %>% fmt_number( columns = vars(mass), decimals = 0 ) %>% cols_align( align = "center", columns = vars(species, mass, height) ) ``` ] ] .smallest[ .pull-right[ .image-tall[ 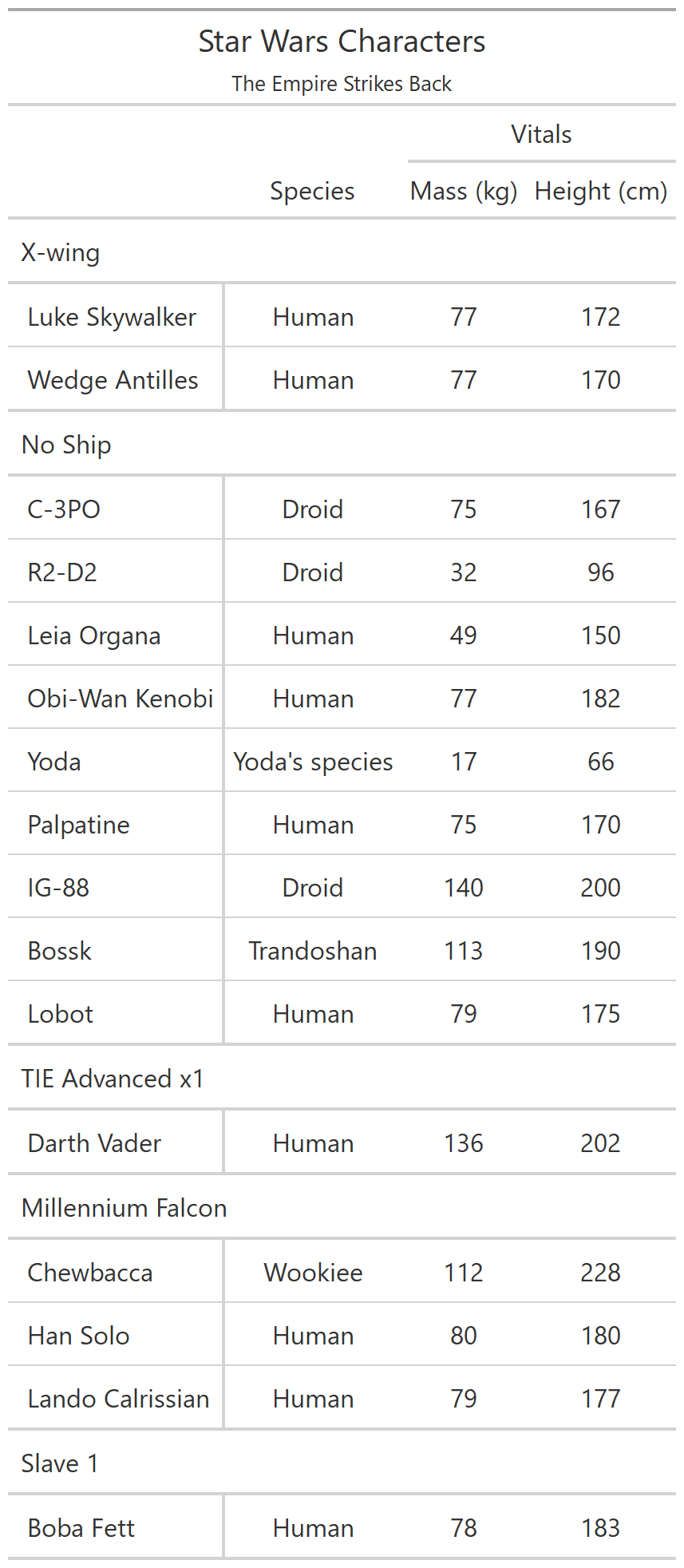<!-- --> ] ] ] --- count: false .pull-left[ ## Reorder Groups .small[ ```r tes_chars %>% group_by(starships) %>% gt(rowname_col = "name") %>% tab_header( title = "Star Wars Characters", subtitle = "The Empire Strikes Back" ) %>% tab_spanner( label = "Vitals", columns = vars(mass, height) ) %>% cols_label( mass = "Mass (kg)", height = "Height (cm)", species = "Species" ) %>% fmt_number( columns = vars(mass), decimals = 0 ) %>% cols_align( align = "center", columns = vars(species, mass, height) ) %>% row_group_order( groups = c("X-wing", "Millennium Falcon") ) ``` ] ] .smallest[ .pull-right[ .image-tall[ 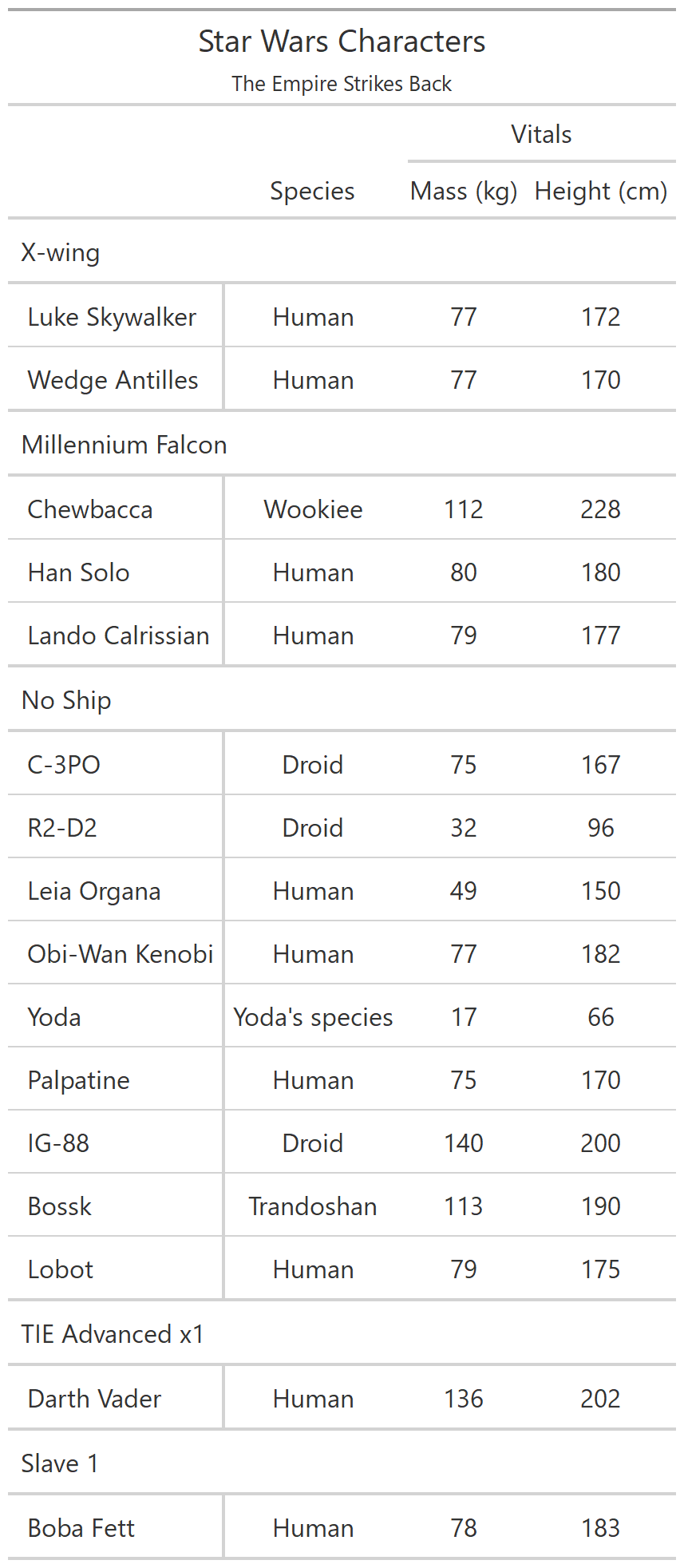<!-- --> ] ] ] --- count: false .pull-left[ ### Big Improvement! .image-full[ 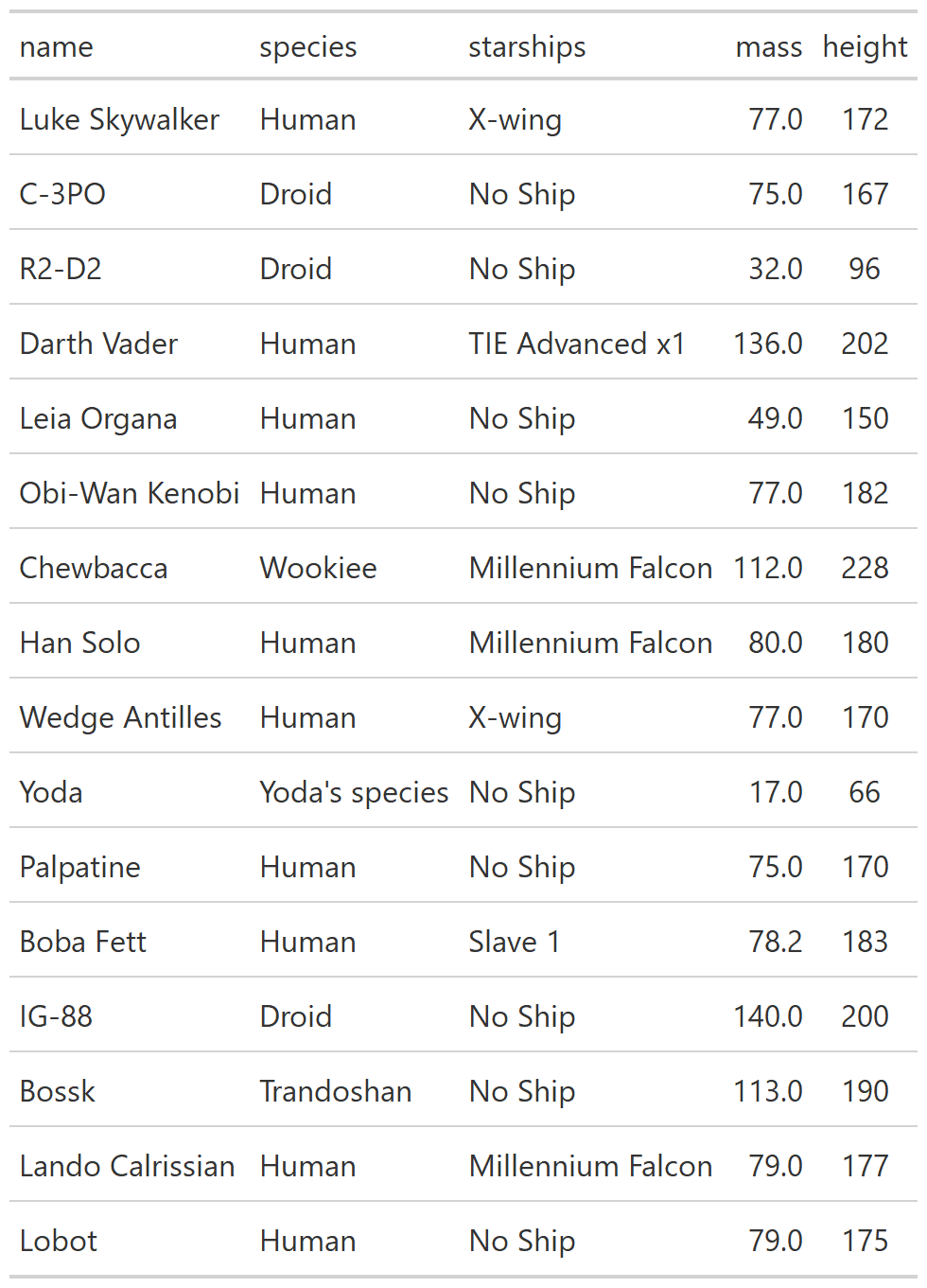<!-- --> ] ] .pull-right[ .image-tall[ 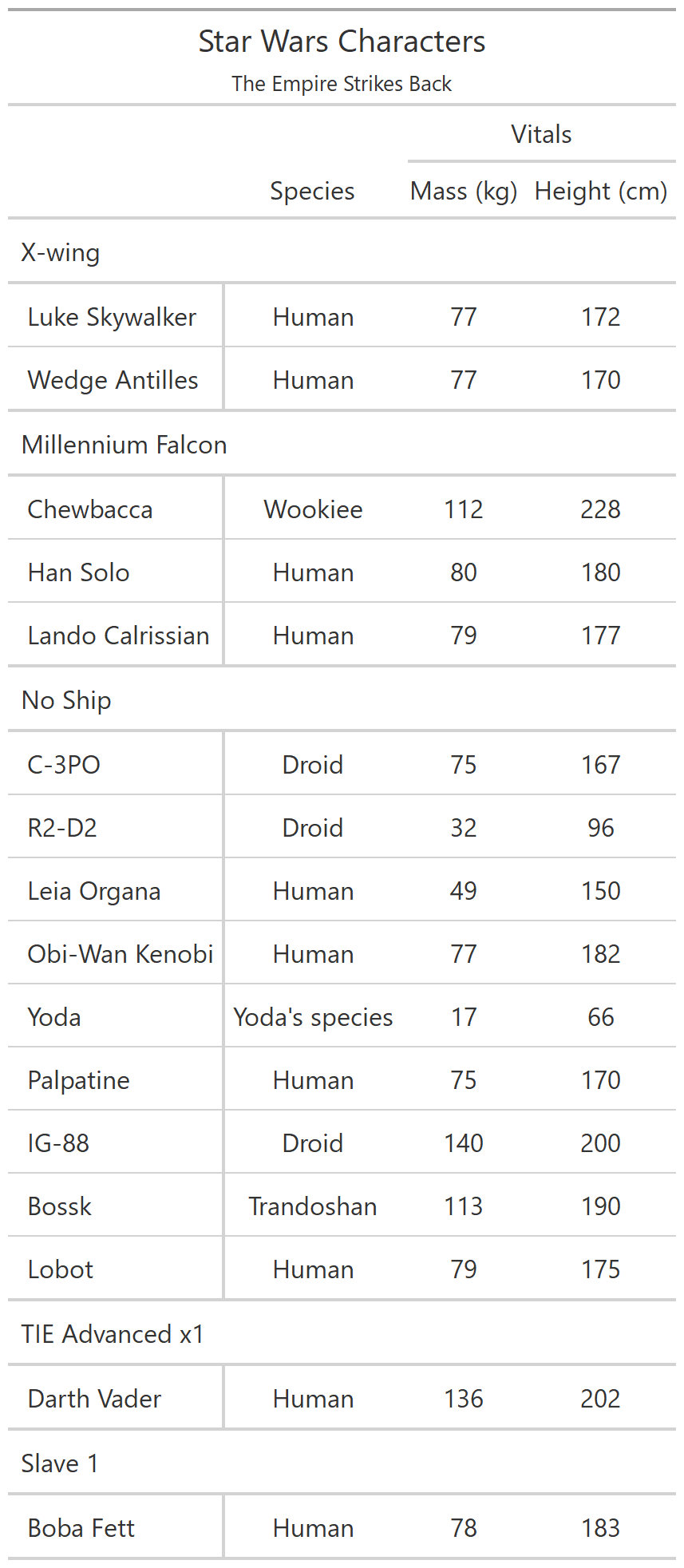<!-- --> ] ] --- # `\(\LaTeX\)` Tables `gt` is a very new package and is somewhat finicky when used in `.pdf` documents. For tables in `\(\LaTeX\)`—as is needed for `.pdf` files—I recommend also looking into the `kableExtra` or `flextable` packages. -- Like `gt`, `kableExtra` and `flextable` allow the construction of complex tables in either HTML or `\(\LaTeX\)` using additive syntax similar to `ggplot2` and `dplyr`. `flextable` is also great for Word tables. -- If you want to edit `\(\LaTeX\)` documents, you can do it in R using Sweave documents (.Rnw). Alternatively, you may want to work in a dedicated `\(\LaTeX\)` editor. I recommend [Overleaf](http://www.overleaf.com) for this purpose. -- RMarkdown has support for a fair amount of basic `\(\LaTeX\)` syntax if you aren't trying to get too fancy! --- .pull-left[ # `flextable` This is a table produced by `flextable` in Word format--including the embedded density images!<sup>1</sup> Look into `flextable` if you'll be working in Word or want a table package that handles just about every format. .footnote[[1] [Embedding summary plots is a bit complicated and requires list columns (see here)](https://ardata-fr.github.io/flextable-book/cell-content-1.html#mini-charts).] ] .pull-right[ .image-full[  ] ] --- # `modelsummary` The `modelsummary` package combines `broom`, `gt`, `flextable`, and `kableExtra` to produce tabular summaries of almost any model fit in R. An advantage of this package is that it can produce output in every common format: HTML, Markdown, `\(\LaTeX\)`, raw text, and even images (`.png` or `.jpg`). ```r library(modelsummary) ``` Its key function is `msummary()` or `modelsummary()` which creates summary tables of models. You can then build on it using `gt`, `flextable`, or `kableExtra` functions, depending on the selected output format. --- .pull-left[ ## `msummary` Like `pander()`, `msummary()` takes a model as an object to make a table. .small[ ```r mod_1 <- lm(mpg ~ wt, data = mtcars) msummary(mod_1) ``` ] Note default `modelsummary` look like `pander` tables because they use Markdown. ] .pull-right[ .small[ <table class="table" style="width: auto !important; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> </th> <th style="text-align:center;"> Model 1 </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> (Intercept) </td> <td style="text-align:center;"> 37.285 </td> </tr> <tr> <td style="text-align:left;"> </td> <td style="text-align:center;"> (1.878) </td> </tr> <tr> <td style="text-align:left;"> wt </td> <td style="text-align:center;"> -5.344 </td> </tr> <tr> <td style="text-align:left;box-shadow: 0px 1px"> </td> <td style="text-align:center;box-shadow: 0px 1px"> (0.559) </td> </tr> <tr> <td style="text-align:left;"> Num.Obs. </td> <td style="text-align:center;"> 32 </td> </tr> <tr> <td style="text-align:left;"> R2 </td> <td style="text-align:center;"> 0.753 </td> </tr> <tr> <td style="text-align:left;"> R2 Adj. </td> <td style="text-align:center;"> 0.745 </td> </tr> <tr> <td style="text-align:left;"> AIC </td> <td style="text-align:center;"> 166.0 </td> </tr> <tr> <td style="text-align:left;"> BIC </td> <td style="text-align:center;"> 170.4 </td> </tr> <tr> <td style="text-align:left;"> Log.Lik. </td> <td style="text-align:center;"> -80.015 </td> </tr> <tr> <td style="text-align:left;"> F </td> <td style="text-align:center;"> 91.375 </td> </tr> </tbody> </table> ] ] --- .pull-left[ ## `msummary` You can present multiple models in `msummary` using named lists: .small[ ```r mod_1 <- lm(mpg ~ wt, data = mtcars) mod_2 <- lm(mpg ~ hp + wt, data = mtcars) mod_3 <- lm(mpg ~ hp + wt + factor(am), data = mtcars) model_list <- list("Model 1" = mod_1, "Model 2" = mod_2, "Model 3" = mod_3) msummary(model_list) ``` ] This allows you to produce the common (and often bad) journal format where one starts with a nonsensical "naive model" then works up to the "full model" justified by the front end of the paper. ] .pull-right[ .small[ <table class="table" style="width: auto !important; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> </th> <th style="text-align:center;"> Model 1 </th> <th style="text-align:center;"> Model 2 </th> <th style="text-align:center;"> Model 3 </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> (Intercept) </td> <td style="text-align:center;"> 37.285 </td> <td style="text-align:center;"> 37.227 </td> <td style="text-align:center;"> 34.003 </td> </tr> <tr> <td style="text-align:left;"> </td> <td style="text-align:center;"> (1.878) </td> <td style="text-align:center;"> (1.599) </td> <td style="text-align:center;"> (2.643) </td> </tr> <tr> <td style="text-align:left;"> wt </td> <td style="text-align:center;"> -5.344 </td> <td style="text-align:center;"> -3.878 </td> <td style="text-align:center;"> -2.879 </td> </tr> <tr> <td style="text-align:left;"> </td> <td style="text-align:center;"> (0.559) </td> <td style="text-align:center;"> (0.633) </td> <td style="text-align:center;"> (0.905) </td> </tr> <tr> <td style="text-align:left;"> hp </td> <td style="text-align:center;"> </td> <td style="text-align:center;"> -0.032 </td> <td style="text-align:center;"> -0.037 </td> </tr> <tr> <td style="text-align:left;"> </td> <td style="text-align:center;"> </td> <td style="text-align:center;"> (0.009) </td> <td style="text-align:center;"> (0.010) </td> </tr> <tr> <td style="text-align:left;"> factor(am)1 </td> <td style="text-align:center;"> </td> <td style="text-align:center;"> </td> <td style="text-align:center;"> 2.084 </td> </tr> <tr> <td style="text-align:left;box-shadow: 0px 1px"> </td> <td style="text-align:center;box-shadow: 0px 1px"> </td> <td style="text-align:center;box-shadow: 0px 1px"> </td> <td style="text-align:center;box-shadow: 0px 1px"> (1.376) </td> </tr> <tr> <td style="text-align:left;"> Num.Obs. </td> <td style="text-align:center;"> 32 </td> <td style="text-align:center;"> 32 </td> <td style="text-align:center;"> 32 </td> </tr> <tr> <td style="text-align:left;"> R2 </td> <td style="text-align:center;"> 0.753 </td> <td style="text-align:center;"> 0.827 </td> <td style="text-align:center;"> 0.840 </td> </tr> <tr> <td style="text-align:left;"> R2 Adj. </td> <td style="text-align:center;"> 0.745 </td> <td style="text-align:center;"> 0.815 </td> <td style="text-align:center;"> 0.823 </td> </tr> <tr> <td style="text-align:left;"> AIC </td> <td style="text-align:center;"> 166.0 </td> <td style="text-align:center;"> 156.7 </td> <td style="text-align:center;"> 156.1 </td> </tr> <tr> <td style="text-align:left;"> BIC </td> <td style="text-align:center;"> 170.4 </td> <td style="text-align:center;"> 162.5 </td> <td style="text-align:center;"> 163.5 </td> </tr> <tr> <td style="text-align:left;"> Log.Lik. </td> <td style="text-align:center;"> -80.015 </td> <td style="text-align:center;"> -74.326 </td> <td style="text-align:center;"> -73.067 </td> </tr> <tr> <td style="text-align:left;"> F </td> <td style="text-align:center;"> 91.375 </td> <td style="text-align:center;"> 69.211 </td> <td style="text-align:center;"> 48.960 </td> </tr> </tbody> </table> ] ] --- # PDF Output `output = "latex"` produces `kableExtra` based output well-suited to PDFs.<sup>1</sup> ```r msummary(model_list, output = "latex") ``` .image-shorter[ 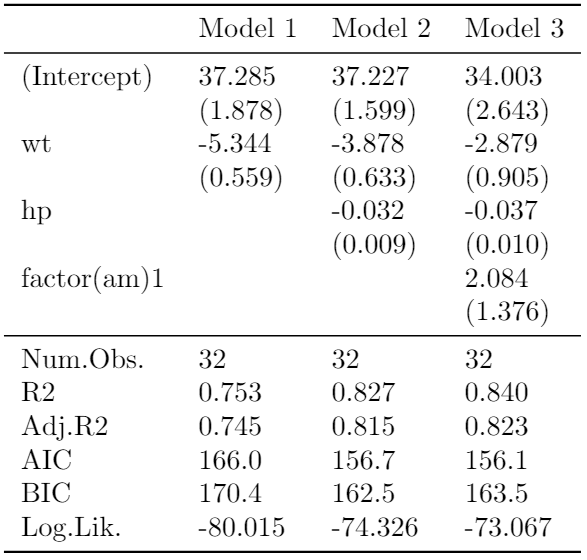 ] For customization, I recommend referring to [`modelsummary`'s documentation](https://vincentarelbundock.github.io/modelsummary/articles/customization.html). --- # Saving a `modelsummary` .image-short[ ```r msummary(model_list, output = "ex_table.png") ``` ] To save a `modelsummary` object as a file, just provide a path to the `output =` argument. Specifying a suffix (e.g., `.png` or `.pdf`) will control the output format. --- # `modelsummary` and `gt` .pull-left[ You can select `gt` output to enable modifying summaries with `gt` functions. .small[ ```r msummary(model_list, output = "gt") %>% tab_header( title = "Table 1. Linear Models", subtitle = "DV: Miles per Gallon" ) ``` ] Note that `gt`'s support for PDF output is immature--this format is better for HTML or image output. ] .pull-right[ .image-full[ 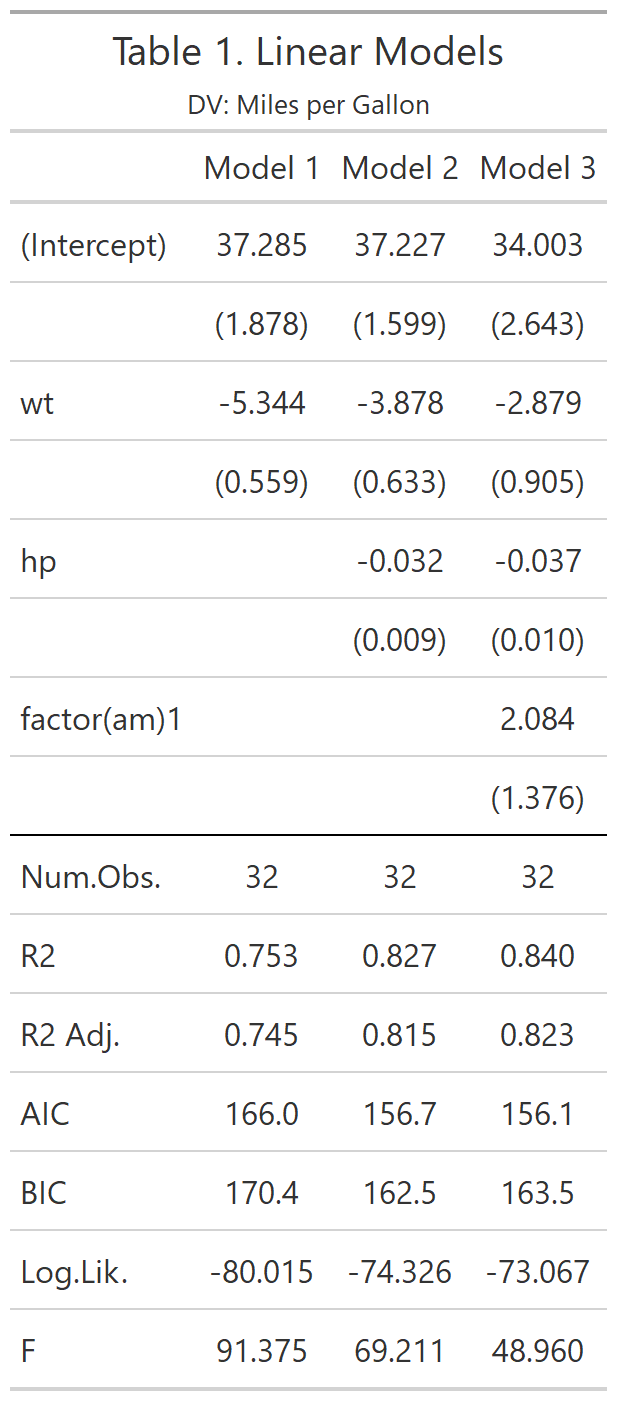<!-- --> ] ] --- # `gtsummary` The `gtsummary` package is similar to `modelsummary` in that it takes advantage of `broom`, `gt`, and `kableExtra` to provide a flexible table-making framework. While `gtsummary` can also produce model tables like `modelsummary`, it also produces descriptive statistic tables for dataframes.<sup>1</sup> ```r library(gtsummary) ``` .footnote[[1] I prefer `modelsummary`'s syntax (or manual table building) for most model tables.] --- .pull-left[ ## `tbl_summary()` By default, `gtsummary` tables provide: * Frequencies for categorical and binary variables * Quantiles of the form "50% (25%, 75%)" for continuous variables * Sample size ```r mtcars %>% select(1:9) %>% tbl_summary() ``` ] .pull-right[ .smaller[ <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #wqyicfvrtk .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #wqyicfvrtk .gt_heading { background-color: #FFFFFF; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #wqyicfvrtk .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #wqyicfvrtk .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 4px; border-top-color: #FFFFFF; border-top-width: 0; } #wqyicfvrtk .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #wqyicfvrtk .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #wqyicfvrtk .gt_col_heading { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #wqyicfvrtk .gt_column_spanner_outer { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #wqyicfvrtk .gt_column_spanner_outer:first-child { padding-left: 0; } #wqyicfvrtk .gt_column_spanner_outer:last-child { padding-right: 0; } #wqyicfvrtk .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; overflow-x: hidden; display: inline-block; width: 100%; } #wqyicfvrtk .gt_group_heading { padding: 8px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; } #wqyicfvrtk .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #wqyicfvrtk .gt_from_md > :first-child { margin-top: 0; } #wqyicfvrtk .gt_from_md > :last-child { margin-bottom: 0; } #wqyicfvrtk .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #wqyicfvrtk .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 12px; } #wqyicfvrtk .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #wqyicfvrtk .gt_first_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; } #wqyicfvrtk .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #wqyicfvrtk .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #wqyicfvrtk .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #wqyicfvrtk .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #wqyicfvrtk .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #wqyicfvrtk .gt_footnote { margin: 0px; font-size: 90%; padding: 4px; } #wqyicfvrtk .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #wqyicfvrtk .gt_sourcenote { font-size: 90%; padding: 4px; } #wqyicfvrtk .gt_left { text-align: left; } #wqyicfvrtk .gt_center { text-align: center; } #wqyicfvrtk .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #wqyicfvrtk .gt_font_normal { font-weight: normal; } #wqyicfvrtk .gt_font_bold { font-weight: bold; } #wqyicfvrtk .gt_font_italic { font-style: italic; } #wqyicfvrtk .gt_super { font-size: 65%; } #wqyicfvrtk .gt_footnote_marks { font-style: italic; font-size: 65%; } </style> <div id="wqyicfvrtk" style="overflow-x:auto;overflow-y:auto;width:auto;height:auto;"><table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1"><strong>Characteristic</strong></th> <th class="gt_col_heading gt_columns_bottom_border gt_center" rowspan="1" colspan="1"><strong>N = 32</strong><sup class="gt_footnote_marks">1</sup></th> </tr> </thead> <tbody class="gt_table_body"> <tr> <td class="gt_row gt_left">mpg</td> <td class="gt_row gt_center">19.2 (15.4, 22.8)</td> </tr> <tr> <td class="gt_row gt_left">cyl</td> <td class="gt_row gt_center"></td> </tr> <tr> <td class="gt_row gt_left" style="text-align: left; text-indent: 10px;">4</td> <td class="gt_row gt_center">11 (34%)</td> </tr> <tr> <td class="gt_row gt_left" style="text-align: left; text-indent: 10px;">6</td> <td class="gt_row gt_center">7 (22%)</td> </tr> <tr> <td class="gt_row gt_left" style="text-align: left; text-indent: 10px;">8</td> <td class="gt_row gt_center">14 (44%)</td> </tr> <tr> <td class="gt_row gt_left">disp</td> <td class="gt_row gt_center">196 (121, 326)</td> </tr> <tr> <td class="gt_row gt_left">hp</td> <td class="gt_row gt_center">123 (96, 180)</td> </tr> <tr> <td class="gt_row gt_left">drat</td> <td class="gt_row gt_center">3.70 (3.08, 3.92)</td> </tr> <tr> <td class="gt_row gt_left">wt</td> <td class="gt_row gt_center">3.33 (2.58, 3.61)</td> </tr> <tr> <td class="gt_row gt_left">qsec</td> <td class="gt_row gt_center">17.71 (16.89, 18.90)</td> </tr> <tr> <td class="gt_row gt_left">vs</td> <td class="gt_row gt_center">14 (44%)</td> </tr> <tr> <td class="gt_row gt_left">am</td> <td class="gt_row gt_center">13 (41%)</td> </tr> </tbody> <tfoot> <tr class="gt_footnotes"> <td colspan="2"> <p class="gt_footnote"> <sup class="gt_footnote_marks"> <em>1</em> </sup> Median (IQR); n (%) <br /> </p> </td> </tr> </tfoot> </table></div> ] ] --- .pull-left[ ## Grouping You can provide a `by = ` argument to do grouped descriptives. ```r mtcars %>% select(1:9) %>% tbl_summary(by = "am") ``` ] .pull-right[ .smaller[ <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #sqketivvan .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #sqketivvan .gt_heading { background-color: #FFFFFF; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #sqketivvan .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #sqketivvan .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 4px; border-top-color: #FFFFFF; border-top-width: 0; } #sqketivvan .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #sqketivvan .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #sqketivvan .gt_col_heading { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #sqketivvan .gt_column_spanner_outer { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #sqketivvan .gt_column_spanner_outer:first-child { padding-left: 0; } #sqketivvan .gt_column_spanner_outer:last-child { padding-right: 0; } #sqketivvan .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; overflow-x: hidden; display: inline-block; width: 100%; } #sqketivvan .gt_group_heading { padding: 8px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; } #sqketivvan .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #sqketivvan .gt_from_md > :first-child { margin-top: 0; } #sqketivvan .gt_from_md > :last-child { margin-bottom: 0; } #sqketivvan .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #sqketivvan .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 12px; } #sqketivvan .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #sqketivvan .gt_first_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; } #sqketivvan .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #sqketivvan .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #sqketivvan .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #sqketivvan .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #sqketivvan .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #sqketivvan .gt_footnote { margin: 0px; font-size: 90%; padding: 4px; } #sqketivvan .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #sqketivvan .gt_sourcenote { font-size: 90%; padding: 4px; } #sqketivvan .gt_left { text-align: left; } #sqketivvan .gt_center { text-align: center; } #sqketivvan .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #sqketivvan .gt_font_normal { font-weight: normal; } #sqketivvan .gt_font_bold { font-weight: bold; } #sqketivvan .gt_font_italic { font-style: italic; } #sqketivvan .gt_super { font-size: 65%; } #sqketivvan .gt_footnote_marks { font-style: italic; font-size: 65%; } </style> <div id="sqketivvan" style="overflow-x:auto;overflow-y:auto;width:auto;height:auto;"><table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_left" rowspan="1" colspan="1"><strong>Characteristic</strong></th> <th class="gt_col_heading gt_columns_bottom_border gt_center" rowspan="1" colspan="1"><strong>0</strong>, N = 19<sup class="gt_footnote_marks">1</sup></th> <th class="gt_col_heading gt_columns_bottom_border gt_center" rowspan="1" colspan="1"><strong>1</strong>, N = 13<sup class="gt_footnote_marks">1</sup></th> </tr> </thead> <tbody class="gt_table_body"> <tr> <td class="gt_row gt_left">mpg</td> <td class="gt_row gt_center">17.3 (14.9, 19.2)</td> <td class="gt_row gt_center">22.8 (21.0, 30.4)</td> </tr> <tr> <td class="gt_row gt_left">cyl</td> <td class="gt_row gt_center"></td> <td class="gt_row gt_center"></td> </tr> <tr> <td class="gt_row gt_left" style="text-align: left; text-indent: 10px;">4</td> <td class="gt_row gt_center">3 (16%)</td> <td class="gt_row gt_center">8 (62%)</td> </tr> <tr> <td class="gt_row gt_left" style="text-align: left; text-indent: 10px;">6</td> <td class="gt_row gt_center">4 (21%)</td> <td class="gt_row gt_center">3 (23%)</td> </tr> <tr> <td class="gt_row gt_left" style="text-align: left; text-indent: 10px;">8</td> <td class="gt_row gt_center">12 (63%)</td> <td class="gt_row gt_center">2 (15%)</td> </tr> <tr> <td class="gt_row gt_left">disp</td> <td class="gt_row gt_center">276 (196, 360)</td> <td class="gt_row gt_center">120 (79, 160)</td> </tr> <tr> <td class="gt_row gt_left">hp</td> <td class="gt_row gt_center">175 (116, 192)</td> <td class="gt_row gt_center">109 (66, 113)</td> </tr> <tr> <td class="gt_row gt_left">drat</td> <td class="gt_row gt_center">3.15 (3.07, 3.70)</td> <td class="gt_row gt_center">4.08 (3.85, 4.22)</td> </tr> <tr> <td class="gt_row gt_left">wt</td> <td class="gt_row gt_center">3.52 (3.44, 3.84)</td> <td class="gt_row gt_center">2.32 (1.94, 2.78)</td> </tr> <tr> <td class="gt_row gt_left">qsec</td> <td class="gt_row gt_center">17.82 (17.18, 19.17)</td> <td class="gt_row gt_center">17.02 (16.46, 18.61)</td> </tr> <tr> <td class="gt_row gt_left">vs</td> <td class="gt_row gt_center">7 (37%)</td> <td class="gt_row gt_center">7 (54%)</td> </tr> </tbody> <tfoot> <tr class="gt_footnotes"> <td colspan="3"> <p class="gt_footnote"> <sup class="gt_footnote_marks"> <em>1</em> </sup> Median (IQR); n (%) <br /> </p> </td> </tr> </tfoot> </table></div> ] ] --- .pull-left[ ## Adding `gt` If you select `gt` output, you can dress it up with `gt` functions. .small[ ```r mtcars %>% select(1:9) %>% tbl_summary(by = "am") %>% as_gt() %>% tab_spanner( label = "Transmission", columns = starts_with("stat_") ) %>% tab_header( title = "Motor Trend Cars", subtitle = "Descriptive Statistics" ) ``` ] `starts\_with("stat\_")` here selects the statistic columns created by `tbl_summary()`. ] .pull-right[ .smaller[ 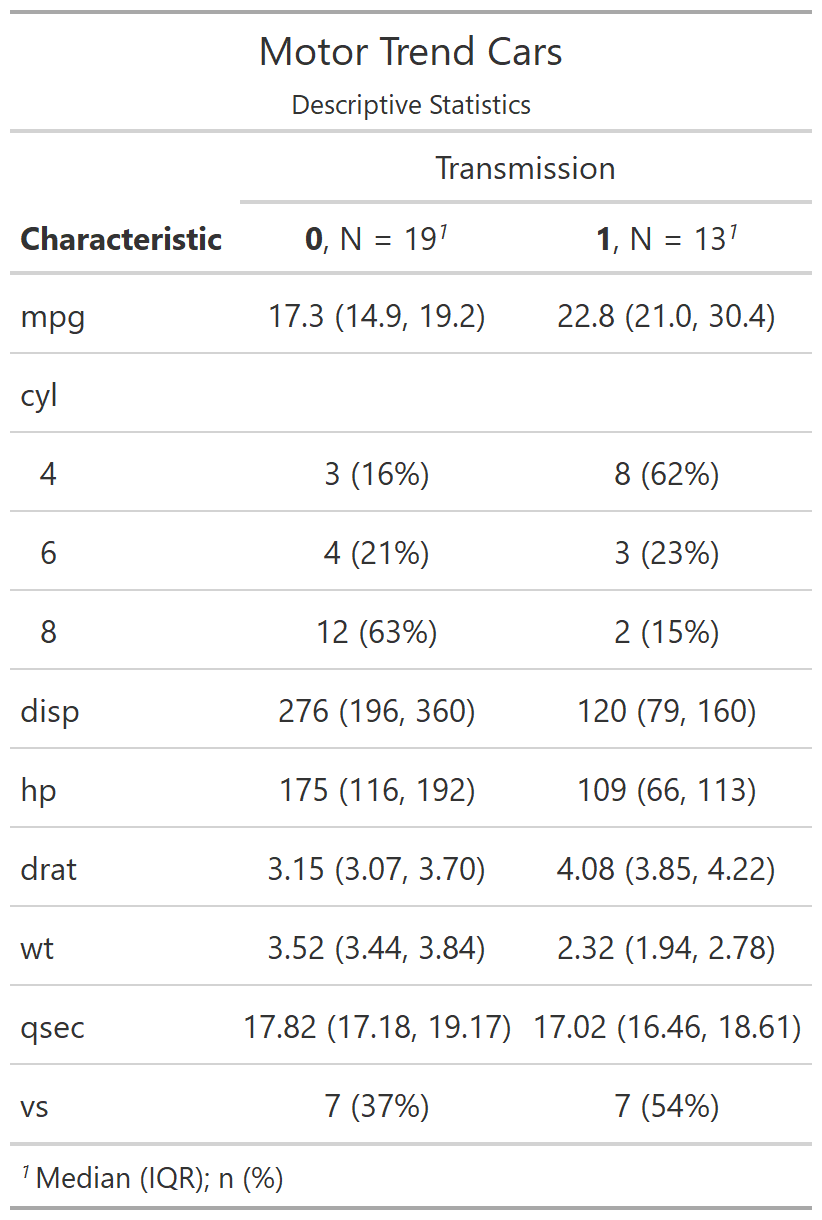<!-- --> ] ] --- # Bonus: `corrplot` The `corrplot` package has functions for displaying correlograms. These make interpreting the correlations between variables in a data set easier than conventional correlation tables. The first argument is a call to `cor()`, the base R function for generating a correlation matrix. [See the vignette for customization options.](https://cran.r-project.org/web/packages/corrplot/vignettes/corrplot-intro.html) ```r library(corrplot) corrplot( cor(mtcars), addCoef.col = "white", addCoefasPercent=T, type="upper", order="AOE") ``` --- ## Correlogram <!-- --> --- class: inverse # Reproducible Research --- # Why Reproducibility? Reproducibility is not *replication*. * **Replication** is running a new study to show if and how results of a prior study hold. * **Reproducibility** is about rerunning *the same study* and getting the *same results*. -- Reproducible studies can still be *wrong*... and in fact reproducibility makes proving a study wrong *much easier*. -- Reproducibility means: * Transparent research practices. * Minimal barriers to verifying your results. -- *Any study that isn't reproducible can be trusted only on faith.* --- # Reproducibility Definitions Reproducibility comes in three forms (Stodden 2014): -- 1. **Empirical:** Repeatability in data collection. -- 2. **Statistical:** Verification with alternate methods of inference. -- 3. **Computational:** Reproducibility in cleaning, organizing, and presenting data and results. -- R is particularly well suited to enabling **computational reproducibility**.<sup>1</sup> .footnote[[1] Python is equally well suited. Julia is an option as well.] -- They will not fix flawed research design, nor offer a remedy for improper application of statistical methods. Those are the difficult, non-automatable things you want skills in. --- ## Computational Reproducibility Elements of computational reproducibility: -- * Shared data + Researchers need your original data to verify and replicate your work. -- * Shared code + Your code must be shared to make decisions transparent. -- * Documentation + The operation of code should be either self-documenting or have written descriptions to make its use clear. -- * **Version Control** + Documents the research process. + Prevents losing work and facilitates sharing. --- ## Levels of Reproducibility For academic papers, degrees of reproducibility vary: 0. "Read the article" -- 1. Shared data with documentation -- 2. Shared data and all code -- 3. **Interactive document** -- 4. **Research compendium** -- 5. Docker compendium: Self-contained ecosystem --- ## Interactive Documents **Interactive documents**—like R Markdown docs—combine code and text together into a self-contained document. * Load and process data * Run models * Generate tables and plots in-line with text * In-text values automatically filled in -- Interactive documents allow a reader to examine your computational methods within the document itself; in effect, they are self-documenting. -- By re-running the code, they reproduce your results on demand. -- Common Platforms: * **R:** R Markdown ([an example of mine](https://clanfear.github.io/birthtiming/inst/paper/paper.html)) * **Python:** Jupyter Notebooks --- ## Research Compendia A **research compendium** is a portable, reproducible distribution of an article or other project. -- Research compendia feature: * An interactive document as the foundation * Files organized in a recognizable structure (e.g. an R package) * Clear separation of data, method, and output. *Data are read only*. * A well-documented or even *preserved* computational environment (e.g. Docker) -- `rrtools` by UW's [Ben Markwick](https://github.com/benmarwick) provides a simplified workflow to accomplish this in R. [Here is an example compendium of mine.](https://github.com/clanfear/birthtiming) --- ## Bookdown [`bookdown`](https://bookdown.org/yihui/bookdown/)—which is integrated into `rrtools`—can generate documents in the proper format for articles, theses, books, or dissertations. -- `bookdown` provides an accessible alternative to writing `\(\LaTeX\)` for typesetting and reference management. -- You can integrate citations and automate reference page generation using bibtex files (such as produced by Zotero). -- `bookdown` supports `.html` output for ease and speed and also renders `.pdf` files through `\(\LaTeX\)` for publication-ready documents. -- For University of Washington theses and dissertations, consider Ben Marwick's [`huskydown` package](https://github.com/benmarwick/huskydown) which uses Markdown but renders via a UW approved `\(\LaTeX\)` template. --- class: inverse # Best Practices ## Organization and Portability --- # Organization Systems Organizing research projects is something you either do accidentally—and badly—or purposefully with some upfront labor. -- Uniform organization makes switching between or revisiting projects easier. -- I suggest something like the following: .pull-left[ ``` project/ readme.md data/ derived/ processed_data.RData raw/ core_data.csv docs/ paper.Rmd syntax/ functions.R models.R ``` ] .pull-right[ 1. There is a clear hierarchy * Written content is in `docs` * Code is in `syntax` * Data is in `data` 2. Naming is uniform * All lower case * Words separated by underscores 3. Names are self-descriptive ] --- # Workflow versus Project To summarize Jenny Bryan, [one should separate workflow from projects.](https://www.tidyverse.org/articles/2017/12/workflow-vs-script/) -- .pull-left[ ### Workflow * The software you use to write your code (e.g. RStudio) * The location you store a project * The specific computer you use * The code you ran earlier or typed into your console ] -- .pull-right[ ### Project * The raw data * The code that operates on your raw data * The packages you use * The output files or documents ] -- Projects *should not modify anything outside of the project* nor need to be modified by someone else (or future you) to run. **Projects *should be independent of your workflow*.** --- # Portability For research to be reproducible, it must also be *portable*. Portable software operates *independently of workflow* such as fixed file locations. -- **Do Not:** * Use `setwd()` in scripts or .Rmd files. * Use *absolute paths* except for *fixed, immovable sources* (secure data). + `read_csv("C:/my_project/data/my_data.csv")` * Use `install.packages()` in script or .Rmd files. * Use `rm(list=ls())` anywhere but your console. -- **Do:** * Use RStudio projects (or the [`here` package](https://github.com/jennybc/here_here)) to set directories. * Use *relative paths* to load and save files: + `read_csv("./data/my_data.csv")` * Load all required packages using `library()`. * Clear your workspace when closing RStudio. + Set *Tools > Global Options... > Save workspace...* to **Never** --- # Divide and Conquer Often you do not want to include all code for a project in one `.Rmd` file: * The code takes too long to knit. * The file is so long it is difficult to read. -- There are two ways to deal with this: 1. Use separate `.R` scripts or `.Rmd` files which save results from complicated parts of a project, then load these results in the main `.Rmd` file. + This is good for loading and cleaning large data. + Also for running slow models. -- 2. Use `source()` to run external `.R` scripts when the `.Rmd` knits. + This can be used to run large files that aren't impractically slow. + Also good for loading project-specific functions. --- # The Way of Many Files I find it beneficial to break projects into *many* files: * Scripts with specialized functions. * Scripts to load and clean each set of variables. * Scripts to run each set of models and make tables and plots. * A main .Rmd that runs some or all of these to reproduce the entire project. -- Splitting up a project carries benefits: * Once a portion of the project is done and in its own file, *it is out of your way.* * If you need to make changes, you don't need to search through huge files. * Entire sections of the project can be added or removed quickly (e.g. converted to an appendix of an article) * **It is the only way to build a proper *pipeline* for a project. ** --- # Pipelines Professional researchers and teams design projects as a **pipeline**. -- A **pipeline** is a series of consecutive processing elements (scripts and functions in R). -- Each stage of a pipeline... 1. Has clearly defined inputs and outputs 2. Does not modify its inputs. 3. Produces the exact same output every time it is re-run. -- This means... 1. When you modify one stage, you only need to rerun *subsequent stages*. 2. Different people can work on each stage. 3. Problems are isolated within stages. 4. You can depict your project as a *directed graph* of **dependencies**. --- # Example Pipeline Every stage (oval) has an unambiguous input and output. Everything that precedes a given stage is a **dependency**—something required to run it.  .footnote[Note: [targets` is a great package for managing R research pipelines.](https://docs.ropensci.org/targets/)] --- class: inverse # Tools ### *Some opinionated advice* --- # On Formats Avoid "closed" or commercial software and file formats except where absolutely necessary. -- Use open source software and file formats. -- * It is always better for *science*: + People should be able to explore your research without buying commercial software. + You do not want your research to be inaccessible when software is updated. -- * It is often just *better*. + It is usually updated more quickly + It tends to be more secure + It is rarely abandoned -- **The ideal:** Use software that reads and writes *raw text*. --- # Text Writing and formatting documents are two completely separate jobs. * Write first * Format later * [Markdown](https://en.wikipedia.org/wiki/Markdown) was made for this -- Word processors—like Microsoft Word—try to do both at the same time, usually badly. They waste time by leading you to format instead of writing. -- Find a good modular text editor and learn to use it: * [Atom](https://atom.io/) * [Sublime](https://www.sublimetext.com/) (Commercial) * Emacs * Vim --- class: inverse # Version Control --- # Version Control Version control originates in collaborative software development. **The Idea:** All changes ever made to a piece of software are documented, saved automatically, and revertible. -- Version control allows all decisions ever made in a research project to be documented automatically. -- Version control can: 1. Protect your work from destructive changes 2. Simplify collaboration by merging changes 3. Document design decisions 4. Make your research process transparent --- # Git and GitHub [`git`](https://en.wikipedia.org/wiki/Git) is the dominant platform for version control, and [GitHub](https://github.com/) is a free (and now Microsoft owned) platform for hosting **repositories**. -- **Repositories** are folders on your computer where all changes are tracked by Git. -- Once satisfied with changes, you "commit" them then "push" them to a remote repository that stores your project. -- Others can copy your project ("pull"), and if you permit, make suggestions for changes. -- Constantly committing and pulling changes automatically generates a running "history" that documents the evolution of a project. -- `git` is integrated into RStudio under the *Tools* menu. [It requires some setup.](http://happygitwithr.com/)<sup>1</sup> .footnote[[1] You can also use the [GitHub desktop application](https://desktop.github.com/).] --- # GitHub as a CV Beyond archiving projects and allowing sharing, GitHub also serves as a sort of curriculum vitae for the programmer. -- By allowing others to view your projects, you can display competence in programming and research. -- If you are planning on working in the private sector, an active GitHub profile will give you a leg up on the competition. -- If you are aiming for academia, a GitHub account signals technical competence and an interest in research transparency. --- class: inverse # Wrapping up the Course --- # What You've Learned A lot! * How to get data into R from a variety of formats * How to do "data custodian" work to manipulate and clean data * How to make pretty visualizations * How to automate with loops and functions * How to combine text, calculations, plots, and tables into dynamic R Markdown reports * How to acquire and work with spatial data --- # What Comes Next? * Statistical inference (e.g. more CSSS courses) + Functions for hypothesis testing, hierarchical/mixed effect models, machine learning, survey design, etc. are straightforward to use... once data are clean + Access output by working with list structures (like from regression models) or using `broom` and `ggeffects` * Practice, practice, practice! + Replicate analyses you've done in Excel, SPSS, or Stata + Think about data using `dplyr` verbs, tidy data principles + R Markdown for reproducibility * More advanced projects + Using version control (git) in RStudio + Interactive Shiny web apps + Write your own functions and put them in a package --- # Course Plugs If you... * have no stats background yet - **SOC504: Applied Social Statistics** * have (only) finished SOC506 - **CSSS510: Maximum Likelihood** * want to master visualization - **CSSS569: Visualizing Data** * study events or durations - **CSSS544: Event History Analysis** <sup>1</sup> * want to use network data - **CSSS567: Social Network Analysis** * want to work with spatial data - **CSSS554: Spatial Statistics** * want to work with time series - **CSSS512: Time Series and Panel Data** .footnote[ [1] Also a great maximum likelihood introduction. ] --- class: inverse # Thank you! --- class: inverse count: false # Supplementary Material --- count: false # `sjPlot` `pander` tables are great for basic `rmarkdown` documents, but they're not generally publication ready. The `sjPlot` package produces `html` tables that look more like those you may find in journal articles. ```r library(sjPlot) ``` --- count: false # `sjPlot` Tables `tab_model()` will produce tables for most models. ```r model_1 <- lm(mpg ~ wt, data = mtcars) tab_model(model_1) ``` <table style="border-collapse:collapse; border:none;"> <tr> <th style="border-top: double; text-align:center; font-style:normal; font-weight:bold; padding:0.2cm; text-align:left; "> </th> <th colspan="3" style="border-top: double; text-align:center; font-style:normal; font-weight:bold; padding:0.2cm; ">mpg</th> </tr> <tr> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; text-align:left; ">Predictors</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; ">Estimates</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; ">CI</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; ">p</td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; ">(Intercept)</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">37.29</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">33.45 – 41.12</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "><strong><0.001</strong></td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; ">wt</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">-5.34</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">-6.49 – -4.20</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "><strong><0.001</strong></td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; padding-top:0.1cm; padding-bottom:0.1cm; border-top:1px solid;">Observations</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; padding-top:0.1cm; padding-bottom:0.1cm; text-align:left; border-top:1px solid;" colspan="3">32</td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; padding-top:0.1cm; padding-bottom:0.1cm;">R<sup>2</sup> / R<sup>2</sup> adjusted</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; padding-top:0.1cm; padding-bottom:0.1cm; text-align:left;" colspan="3">0.753 / 0.745</td> </tr> </table> --- count: false # Multi-Model Tables with `sjTable` Often in journal articles you will see a single table that compares multiple models. Typically, authors will start with a simple model on the left, then add variables, until they have their most complex model on the right. The `sjPlot` package makes this easy to do: just give `tab_model()` more models! --- count: false # Multiple `tab_model()` .small[ ```r model_2 <- lm(mpg ~ hp + wt, data = mtcars) model_3 <- lm(mpg ~ hp + wt + factor(am), data = mtcars) tab_model(model_1, model_2, model_3) ``` <table style="border-collapse:collapse; border:none;"> <tr> <th style="border-top: double; text-align:center; font-style:normal; font-weight:bold; padding:0.2cm; text-align:left; "> </th> <th colspan="3" style="border-top: double; text-align:center; font-style:normal; font-weight:bold; padding:0.2cm; ">mpg</th> <th colspan="3" style="border-top: double; text-align:center; font-style:normal; font-weight:bold; padding:0.2cm; ">mpg</th> <th colspan="3" style="border-top: double; text-align:center; font-style:normal; font-weight:bold; padding:0.2cm; ">mpg</th> </tr> <tr> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; text-align:left; ">Predictors</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; ">Estimates</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; ">CI</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; ">p</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; ">Estimates</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; ">CI</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; col7">p</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; col8">Estimates</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; col9">CI</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; 0">p</td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; ">(Intercept)</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">37.29</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">33.45 – 41.12</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "><strong><0.001</strong></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">37.23</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">33.96 – 40.50</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; col7"><strong><0.001</strong></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; col8">34.00</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; col9">28.59 – 39.42</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; 0"><strong><0.001</strong></td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; ">wt</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">-5.34</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">-6.49 – -4.20</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "><strong><0.001</strong></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">-3.88</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">-5.17 – -2.58</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; col7"><strong><0.001</strong></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; col8">-2.88</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; col9">-4.73 – -1.02</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; 0"><strong>0.004</strong></td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; ">hp</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">-0.03</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">-0.05 – -0.01</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; col7"><strong>0.001</strong></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; col8">-0.04</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; col9">-0.06 – -0.02</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; 0"><strong>0.001</strong></td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; ">am [1]</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; col7"></td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; col8">2.08</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; col9">-0.74 – 4.90</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; 0">0.141</td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; padding-top:0.1cm; padding-bottom:0.1cm; border-top:1px solid;">Observations</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; padding-top:0.1cm; padding-bottom:0.1cm; text-align:left; border-top:1px solid;" colspan="3">32</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; padding-top:0.1cm; padding-bottom:0.1cm; text-align:left; border-top:1px solid;" colspan="3">32</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; padding-top:0.1cm; padding-bottom:0.1cm; text-align:left; border-top:1px solid;" colspan="3">32</td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; padding-top:0.1cm; padding-bottom:0.1cm;">R<sup>2</sup> / R<sup>2</sup> adjusted</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; padding-top:0.1cm; padding-bottom:0.1cm; text-align:left;" colspan="3">0.753 / 0.745</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; padding-top:0.1cm; padding-bottom:0.1cm; text-align:left;" colspan="3">0.827 / 0.815</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; padding-top:0.1cm; padding-bottom:0.1cm; text-align:left;" colspan="3">0.840 / 0.823</td> </tr> </table> ] --- count: false # `sjPlot` does a lot more The `sjPlot` package does *a lot* more than just make pretty tables. It is a rabbit hole of *incredibly* powerful and useful functions for displaying descriptive and inferential results. View the [package website](http://www.strengejacke.de/sjPlot/) for extensive documentation. `sjPlot` is a bit more complicated than `ggeffects` but can do just about everything it can do as well; they were written by the same author! `sjPlot` is fairly new but offers a fairly comprehensive solution for `ggplot` based publication-ready social science data visualization. All graphical functions in `sjPlot` are based on `ggplot2`, so it should not take terribly long to figure out. --- count: false # `sjPlot` Example: Likert plots <img src="img/sjPlot_likert.PNG" width="600px" /> --- count: false # `sjPlot` Example: Crosstabs <img src="img/sjPlot_crosstab.PNG" width="500px" />